3GR1
 
 | |
3GMY
 
 | |
3GMV
 
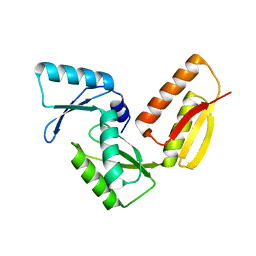
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Apo Form | | Descriptor: | Beta-lactamase inhibitory protein BLIP-I, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
2X61
 
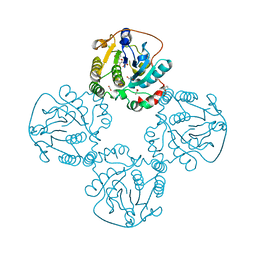 | | Crystal structure of the sialyltransferase CST-II in complex with trisaccharide acceptor and CMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
3HOE
 
 | |
3HOL
 
 | |
3IR6
 
 | | Crystal structure of NarGHI mutant NarG-H49S | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
3IR5
 
 | | Crystal structure of NarGHI mutant NarG-H49C | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
3IR7
 
 | | Crystal structure of NarGHI mutant NarG-R94S | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
3J1X
 
 | | A refined model of the prototypical Salmonella typhimurium T3SS basal body reveals the molecular basis for its assembly | | Descriptor: | Protein PrgH | | Authors: | Sgourakis, N.G, Bergeron, J.R.C, Worrall, L.J, Strynadka, N.C.J, Baker, D. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | A Refined Model of the Prototypical Salmonella SPI-1 T3SS Basal Body Reveals the Molecular Basis for Its Assembly.
Plos Pathog., 9, 2013
|
|
3J83
 
 | |
3J6D
 
 | | Model of the PrgH-PrgK periplasmic rings | | Descriptor: | Pathogenicity 1 island effector protein, Protein PrgH | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-02-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
3J1W
 
 | |
3L7L
 
 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (30 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Lovering, A.L, Strynadka, N.C.J. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LCE
 
 | | Crystal Structure of Oxa-10 Beta-Lactamase Covalently Bound to Cyclobutanone Beta-Lactam Mimic | | Descriptor: | (1S,3S,4S,5S)-7,7-dichloro-3-methoxy-2-thiabicyclo[3.2.0]heptan-6-one-4-carboxylic acid, Beta-lactamase OXA-10, GLYCEROL, ... | | Authors: | Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2010-01-10 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclobutanone Analogues of beta-Lactams Revisited: Insights into Conformational Requirements for Inhibition of Serine- and Metallo-beta-Lactamases.
J.Am.Chem.Soc., 132, 2010
|
|
