7B27
 
 | |
5EFV
 
 | | The host-recognition device of Staphylococcus aureus phage Phi11 | | Descriptor: | FE (III) ION, Phi ETA orf 56-like protein | | Authors: | Koc, C, Kuhner, P, Xia, G, Spinelli, S, Roussel, A, Cambillau, C, Stehle, T. | | Deposit date: | 2015-10-26 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the host-recognition device of Staphylococcus aureus phage 11.
Sci Rep, 6, 2016
|
|
7PA6
 
 | | JC polyomavirus VP1 in complex with scFv 27C11 | | Descriptor: | Major capsid protein VP1, scFv 27C11 antibody heavy chain | | Authors: | Harprecht, C, Stroeh, L.J, Nagel, F, Freytag, J, Stehle, T. | | Deposit date: | 2021-07-29 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of human neutralizing antibodies against JC and BK polyomavirus
To Be Published
|
|
7PA9
 
 | |
7PA8
 
 | |
3MOR
 
 | | Crystal structure of Cathepsin B from Trypanosoma Brucei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin B-like cysteine protease, ... | | Authors: | Cupelli, K, Stehle, T. | | Deposit date: | 2010-04-23 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | In vivo protein crystallization opens new routes in structural biology.
Nat.Methods, 9, 2012
|
|
7QD7
 
 | | TarM(Se)_G117R | | Descriptor: | CHLORIDE ION, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-11-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
7QH9
 
 | | TarM(Se)_G117R-4RboP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TarM(Se)_G117R-4RboP, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-12-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
7QNT
 
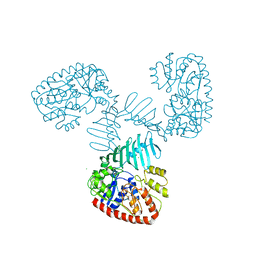 | | TarM(Se) native | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-12-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
3NXG
 
 | | JC polyomavirus VP1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1 | | Authors: | Neu, U, Stroeh, L.J, Stehle, T. | | Deposit date: | 2010-07-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif.
Cell Host Microbe, 8, 2010
|
|
3NXD
 
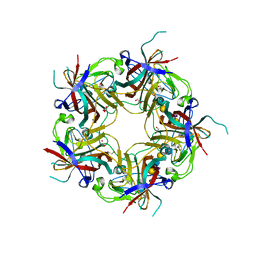 | | JC polyomavirus VP1 in complex with LSTc | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1, ... | | Authors: | Neu, U, Stroeh, L.J, Stehle, T. | | Deposit date: | 2010-07-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif.
Cell Host Microbe, 8, 2010
|
|
3EOY
 
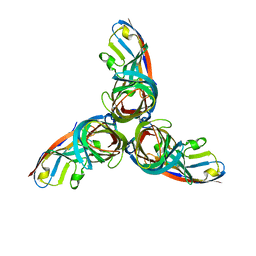 | | Structure of Reovirus sigma1 in Complex with Its Receptor Junctional Adhesion Molecule-A | | Descriptor: | Junctional adhesion molecule A, Outer capsid protein sigma-1 | | Authors: | Kirchner, E, Guglielmi, K.M, Dermody, T.S, Stehle, T. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of reovirus sigma1 in complex with its receptor junctional adhesion molecule-A
Plos Pathog., 4, 2008
|
|
3ZYA
 
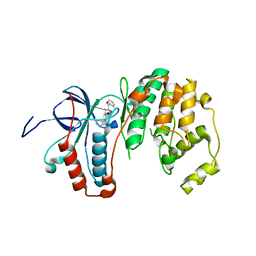 | | Human p38 MAP Kinase in Complex with 2-amino-phenylamino- dibenzosuberone | | Descriptor: | 2-AMINO-PHENYLAMINO-DIBENZOSUBERONE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Romir, J, Koeberle, S.C, Laufer, S.A, Stehle, T. | | Deposit date: | 2011-08-18 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Skepinone-L is a Selective P38 Mitogen-Activated Protein Kinase Inhibitor.
Nat.Chem.Biol., 8, 2011
|
|
4MJ0
 
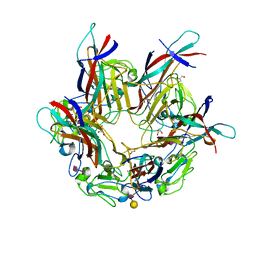 | | BK Polyomavirus VP1 pentamer in complex with GD3 oligosaccharide | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Stroh, L.J, Stehle, T. | | Deposit date: | 2013-09-03 | | Release date: | 2013-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus.
Plos Pathog., 9, 2013
|
|
4MJ1
 
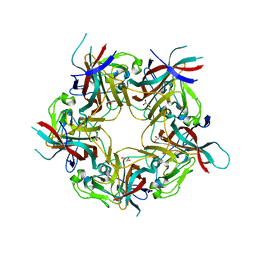 | | unliganded BK Polyomavirus VP1 pentamer | | Descriptor: | CHLORIDE ION, GLYCEROL, VP1 capsid protein | | Authors: | Neu, U, Stroh, L.J, Stehle, T. | | Deposit date: | 2013-09-03 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus.
Plos Pathog., 9, 2013
|
|
4E0U
 
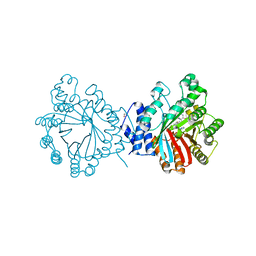 | | Crystal structure of CdpNPT in complex with thiolodiphosphate and (S)-benzodiazependione | | Descriptor: | (3S)-3-(1H-indol-3-ylmethyl)-3,4-dihydro-1H-1,4-benzodiazepine-2,5-dione, 1,2-ETHANEDIOL, Cyclic dipeptide N-prenyltransferase, ... | | Authors: | Schuller, J.M, Zocher, G, Stehle, T. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
4E0T
 
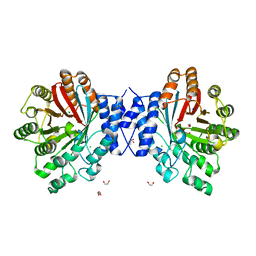 | | Crystal structure of CdpNPT in its unbound state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclic dipeptide N-prenyltransferase, ... | | Authors: | Schuller, J.M, Zocher, G, Stehle, T. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
4EE6
 
 | |
4EQM
 
 | |
4EE8
 
 | |
4EE7
 
 | |
4EPC
 
 | |
4Y7T
 
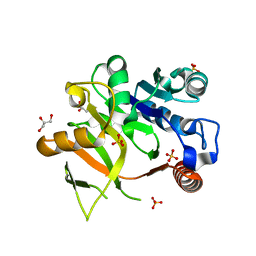 | | Structural analysis of MurU | | Descriptor: | GLYCEROL, Nucleotidyl transferase, SULFATE ION | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
1U8C
 
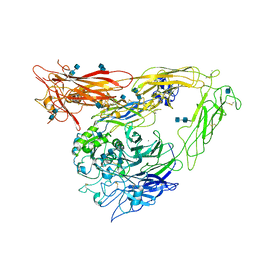 | | A novel adaptation of the integrin PSI domain revealed from its crystal structure | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.P, Stehle, T, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2004-08-05 | | Release date: | 2004-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A novel adaptation of the integrin PSI domain revealed from its crystal structure.
J.Biol.Chem., 279, 2004
|
|
4Y7U
 
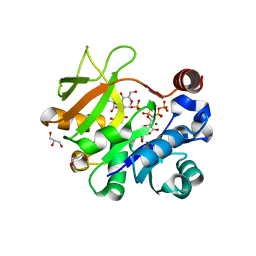 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, GLYCEROL, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
