3DWV
 
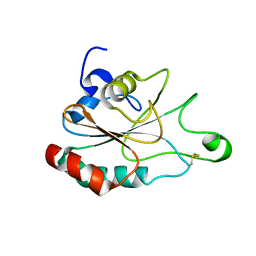 | |
3DEF
 
 | | Crystal structure of Toc33 from Arabidopsis thaliana, dimerization deficient mutant R130A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 protein | | Authors: | Koenig, P, Schleiff, E, Sinning, I, Tews, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | On the significance of Toc-GTPase homodimers
J.Biol.Chem., 283, 2008
|
|
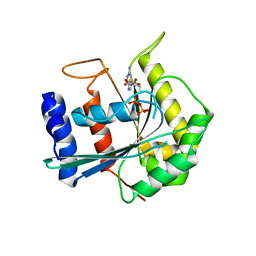
3DEP
 
 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
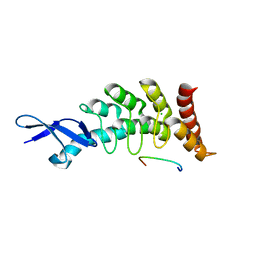
3FEM
 
 | |
2FH5
 
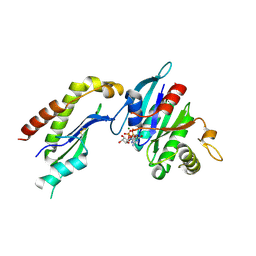 | | The Structure of the Mammalian SRP Receptor | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Signal recognition particle receptor alpha subunit, ... | | Authors: | Schlenker, O, Wild, K, Sinning, I. | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the mammalian signal recognition particle (SRP) receptor as prototype for the interaction of small GTPases with Longin domains.
J.Biol.Chem., 281, 2006
|
|
3DEO
 
 | |
2YHS
 
 | | Structure of the E. coli SRP receptor FtsY | | Descriptor: | 1,2-ETHANEDIOL, CELL DIVISION PROTEIN FTSY | | Authors: | Stjepanovic, G, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2011-05-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Lipids Trigger a Conformational Switch that Regulates Signal Recognition Particle (Srp)-Mediated Protein Targeting.
J.Biol.Chem., 286, 2011
|
|
4GMO
 
 | | Crystal structure of Syo1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GMQ
 
 | | Ribosome-binding domain of Zuo1 | | Descriptor: | Putative ribosome associated protein | | Authors: | Kopp, J, Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GMN
 
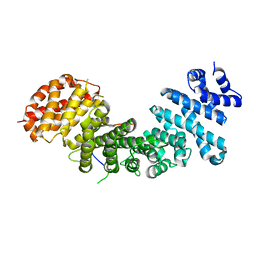 | | Structural basis of Rpl5 recognition by Syo1 | | Descriptor: | 60S ribosomal protein l5-like protein, Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GNI
 
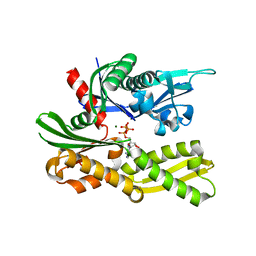 | | Structure of the Ssz1 ATPase bound to ATP and Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative heat shock protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-17 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3BS6
 
 | |
6Y2Z
 
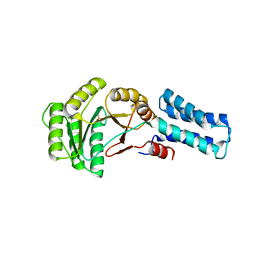 | | NG domain of human SRP54 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
5CK5
 
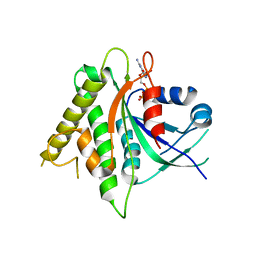 | |
6Y30
 
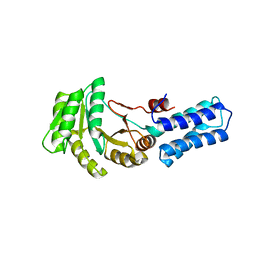 | | NG domain of human SRP54 T115A mutant | | Descriptor: | Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
5CK3
 
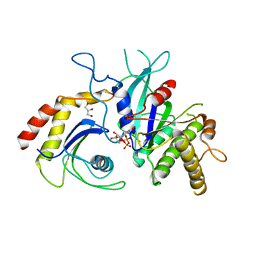 | | Signal recognition particle receptor SRb-GTP/SRX complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jadhav, B.R, Wild, K, Sinning, I. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Switch Cycle of SR beta as Ancestral Eukaryotic GTPase Associated with Secretory Membranes.
Structure, 23, 2015
|
|
5E4X
 
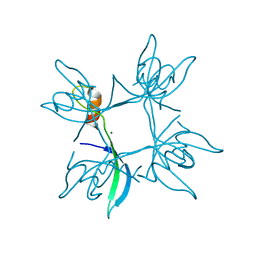 | | Crystal structure of cpSRP43 chromodomain 3 | | Descriptor: | MAGNESIUM ION, Signal recognition particle 43 kDa protein, chloroplastic | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
4ZN4
 
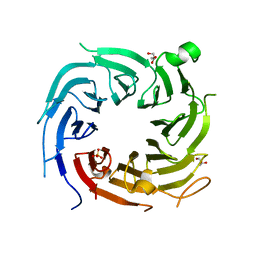 | |
5NZR
 
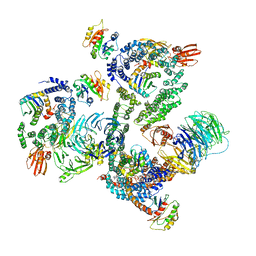 | | The structure of the COPI coat leaf | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5E4W
 
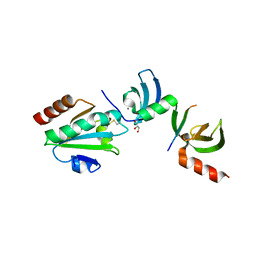 | | Crystal structure of cpSRP43 chromodomains 2 and 3 in complex with the Alb3 tail | | Descriptor: | CALCIUM ION, GLYCEROL, Inner membrane protein ALBINO3, ... | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
5NZU
 
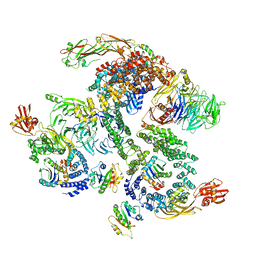 | | The structure of the COPI coat linkage II | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5EM2
 
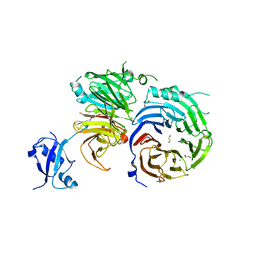 | | Crystal structure of the Erb1-Ytm1 complex | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ribosome biogenesis protein ERB1, ... | | Authors: | Ahmed, Y.L, Sinning, I. | | Deposit date: | 2015-11-05 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Nucleic Acids Res., 44, 2016
|
|
5NZT
 
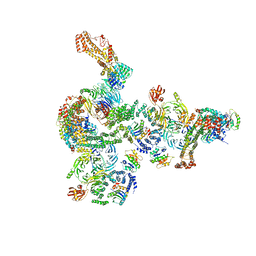 | | The structure of the COPI coat linkage I | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZV
 
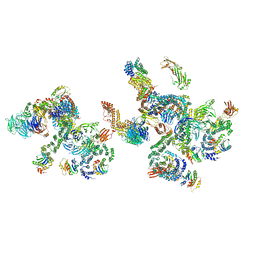 | | The structure of the COPI coat linkage IV | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (17.299999 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5LNS
 
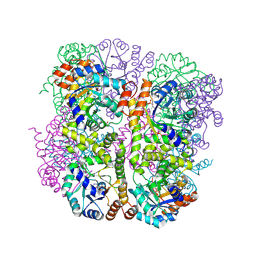 | | Crystal structure of Arabidopsis thaliana Pdx1-R5P complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, RIBULOSE-5-PHOSPHATE | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
