5CMX
 
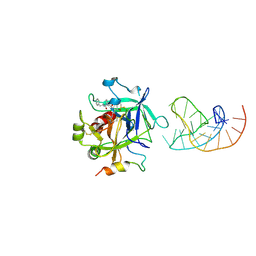 | | X-ray structure of the complex between human alpha thrombin and a duplex/quadruplex 31-mer DNA aptamer | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, RE31, ... | | Authors: | Russo Krauss, I, Pica, A, Napolitano, V, Sica, F. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding.
Nucleic Acids Res., 44, 2016
|
|
1NVG
 
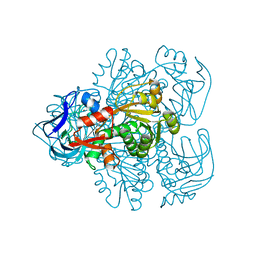 | | N249Y MUTANT OF THE ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS-TETRAGONAL CRYSTAL FORM | | Descriptor: | NAD-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Esposito, L, Bruno, I, Sica, F, Raia, C.A, Giordano, A, Rossi, M, Mazzarella, L, Zagari, A. | | Deposit date: | 2003-02-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural study of a single-point mutant of Sulfolobus solfataricus alcohol dehydrogenase with enhanced activity
Febs Lett., 539, 2003
|
|
1NTO
 
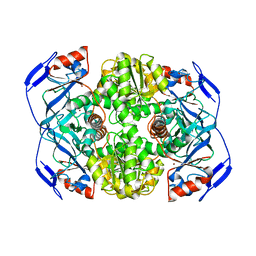 | | N249Y MUTANT OF ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS-MONOCLINIC CRYSTAL FORM | | Descriptor: | NAD-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Esposito, L, Bruno, I, Sica, F, Raia, C.A, Giordano, A, Rossi, M, Mazzarella, L, Zagari, A. | | Deposit date: | 2003-01-30 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural study of a single-point mutant of Sulfolobus solfataricus alcohol dehydrogenase with enhanced activity.
Febs Lett., 539, 2003
|
|
1JVB
 
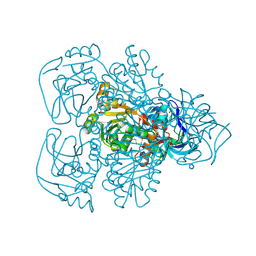 | | ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS | | Descriptor: | NAD(H)-DEPENDENT ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Esposito, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-08-29 | | Release date: | 2002-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the alcohol dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus at 1.85 A resolution.
J.Mol.Biol., 318, 2002
|
|
3QLP
 
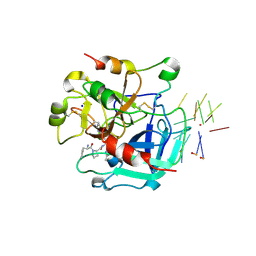 | | X-ray structure of the complex between human alpha thrombin and a modified thrombin binding aptamer (mTBA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2011-02-03 | | Release date: | 2011-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thrombin-aptamer recognition: a revealed ambiguity.
Nucleic Acids Res., 39, 2011
|
|
1R3M
 
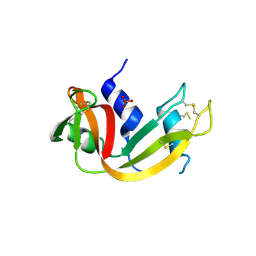 | | Crystal structure of the dimeric unswapped form of bovine seminal ribonuclease | | Descriptor: | PHOSPHATE ION, Ribonuclease, seminal | | Authors: | Berisio, R, Sica, F, De Lorenzo, C, Di Fiore, A, Piccoli, R, Zagari, A, Mazzarella, L. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dimeric unswapped form of bovine seminal ribonuclease
Febs Lett., 554, 2003
|
|
3RID
 
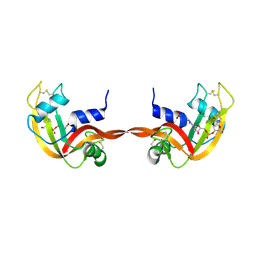 | | X-ray structure of the C-terminal swapped dimer of P114A variant of Ribonuclease A | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE, PHOSPHATE ION, Ribonuclease pancreatic | | Authors: | Merlino, A, Balsamo, A, Mazzarella, L, Sica, F. | | Deposit date: | 2011-04-13 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Chain termini cross-talk in the swapping process of bovine pancreatic ribonuclease.
Biochimie, 94, 2012
|
|
3RH1
 
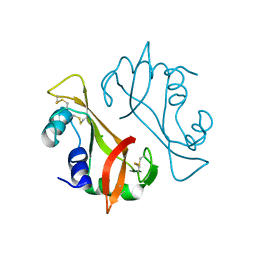 | | X-ray Structure of a cis-proline (P114) to alanine variant of Ribonuclease A | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Merlino, A, Balsamo, A, Mazzarella, L, Sica, F. | | Deposit date: | 2011-04-11 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chain termini cross-talk in the swapping process of bovine pancreatic ribonuclease.
Biochimie, 94, 2012
|
|
8BRC
 
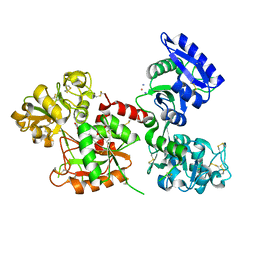 | | Crystal structure of the adduct between human serum transferrin and cisplatin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIA, FE (III) ION, ... | | Authors: | Troisi, R, Galardo, F, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2022-11-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Cisplatin Binding to Human Serum Transferrin: A Crystallographic Study.
Inorg.Chem., 62, 2023
|
|
1KF4
 
 | | Atomic Resolution Structure of RNase A at pH 6.3 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF7
 
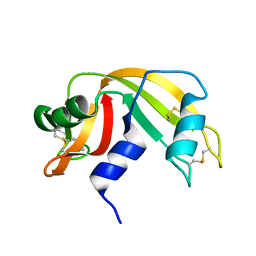 | | Atomic Resolution Structure of RNase A at pH 8.0 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF8
 
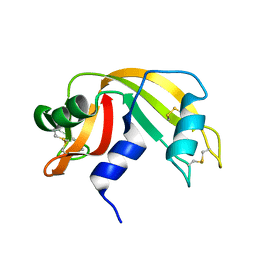 | | Atomic resolution structure of RNase A at pH 8.8 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF3
 
 | | Atomic Resolution Structure of RNase A at pH 5.9 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF5
 
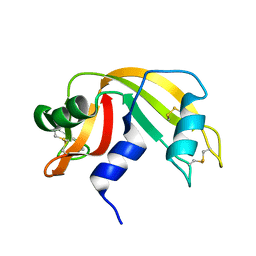 | | Atomic Resolution Structure of RNase A at pH 7.1 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6Z8W
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3G), which contains 1-beta-D-glucopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
6Z8X
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3Leu), which contains leucyl amide in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
6Z8V
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3L), which contains 1-beta-D-lactopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
3GKV
 
 | | X-ray structure of an intermediate along the oxidation pathway of Trematomus bernacchii hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Merlino, A, Vitagliano, L, Sica, F, Vergara, A, Mazzarella, L. | | Deposit date: | 2009-03-11 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Combined crystallographic and spectroscopic analysis of Trematomus bernacchii hemoglobin highlights analogies and differences in the peculiar oxidation pathway of Antarctic fish hemoglobins
Biopolymers, 91, 2009
|
|
3LJ9
 
 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis in complex with sodium azide | | Descriptor: | AZIDE ION, FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
3LJE
 
 | | The X-ray structure of zebrafish RNase5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase5 | | Authors: | Russo Krauss, I, Merlino, A, Coscia, F, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
3LIO
 
 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis (crystal form I) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
3LJF
 
 | | The X-ray structure of iron superoxide dismutase from Pseudoalteromonas haloplanktis (crystal form II) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
4IAU
 
 | | Atomic resolution structure of Geodin, a beta-gamma crystallin from Geodia cydonium | | Descriptor: | Beta-gamma-crystallin, CALCIUM ION, GLYCEROL | | Authors: | Vergara, A, Grassi, M, Sica, F, Mazzarella, L, Merlino, A. | | Deposit date: | 2012-12-07 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A novel interdomain interface in crystallins: structural characterization of the [beta][gamma]-crystallin from Geodia cydonium at 0.99 A resolution
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3LN8
 
 | | The X-ray structure of Zf-RNase-1 from a new crystal form at pH 7.3 | | Descriptor: | HYDROLASE, SULFATE ION | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2010-02-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
3LJD
 
 | | The X-ray structure of zebrafish RNase1 from a new crystal form at pH 4.5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase1 | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
