3HPA
 
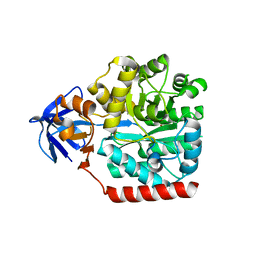 | | Crystal structure of an amidohydrolase gi:44264246 from an evironmental sample of sargasso sea | | Descriptor: | AMIDOHYDROLASE, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The hunt for 8-oxoguanine deaminase.
J.Am.Chem.Soc., 132, 2010
|
|
3ID7
 
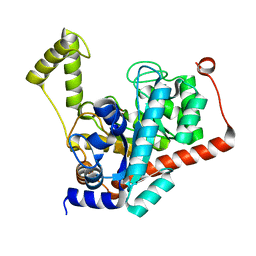 | | Crystal structure of renal dipeptidase from Streptomyces coelicolor A3(2) | | Descriptor: | CHLORIDE ION, Dipeptidase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-07-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3ITC
 
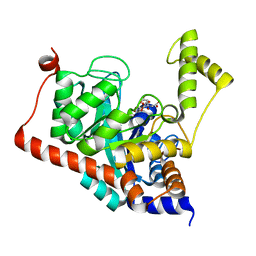 | | Crystal structure of Sco3058 with bound citrate and glycerol | | Descriptor: | CITRIC ACID, GLYCEROL, ZINC ION, ... | | Authors: | Nguyen, T.T, Cummings, J.A, Tsai, C.-L, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase
Biochemistry, 49, 2010
|
|
3K5X
 
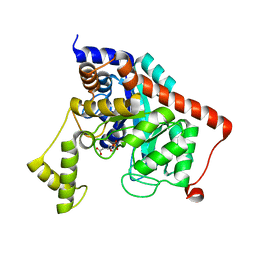 | | Crystal structure of dipeptidase from Streptomics coelicolor complexed with phosphinate pseudodipeptide L-Ala-D-Asp at 1.4A resolution. | | Descriptor: | Dipeptidase, ZINC ION, phosphinate pseudodipeptide L-Ala-D-Asp | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
2AAZ
 
 | | Cryptococcus neoformans thymidylate synthase complexed with substrate and an antifolate | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Finer-Moore, J.S, Anderson, A.C, O'Neil, R.H, Costi, M.P, Ferrari, S, Krucinski, J, Stroud, R.M. | | Deposit date: | 2005-07-14 | | Release date: | 2005-12-06 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of Cryptococcus neoformans thymidylate synthase suggests strategies for using target dynamics for species-specific inhibition.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5SPY
 
 | |
5SQ1
 
 | |
5SQI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5016127255 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQZ
 
 | |
5SQ2
 
 | |
5SQ3
 
 | |
5SQ4
 
 | |
5SPZ
 
 | |
5SQO
 
 | |
5SQ0
 
 | |
5SQX
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5183357278 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-[2-amino-4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[2-amino-4-(cyclopropylcarbamamido)benzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPX
 
 | |
5SQM
 
 | |
5SQ5
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894407 - (R,S) and (S,S) isomers | | Descriptor: | (1R,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQ9
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894420 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SR0
 
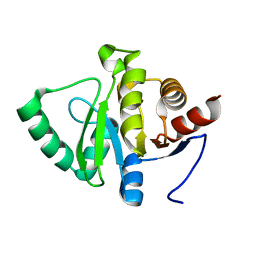 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z3649721459 - (R,S) and (S,R) isomers | | Descriptor: | (1R,2S)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclohexan-1-ol, (1S,2R)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclohexan-1-ol, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQ6
 
 | |
5SQ7
 
 | |
5SQC
 
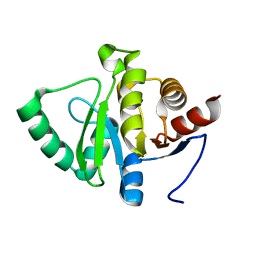 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894388 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQA
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894395 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
