8EUO
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | to be published
To Be Published
|
|
5IRS
 
 | | crystal structure of the proteasomal Rpn13 PRU-domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Proteasomal ubiquitin receptor ADRM1 | | Authors: | Chen, X, Shi, K, Walters, K, Aihara, H. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
7TGR
 
 | | Structure of SARS-CoV-2 main protease in complex with GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Esler, M.A, Shi, K, Aihara, H, Harris, R.S. | | Deposit date: | 2022-01-09 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Gain-of-Signal Assays for Probing Inhibition of SARS-CoV-2 M pro /3CL pro in Living Cells.
Mbio, 13, 2022
|
|
6CH1
 
 | |
6CH3
 
 | |
6CH2
 
 | | Crystal structure of the cytoplasmic domain of FlhA and FliT-FliD complex | | Descriptor: | Flagellar biosynthesis protein FlhA, Flagellar hook-associated protein 2,Flagellar protein FliT, GLYCEROL | | Authors: | Xing, Q, Shi, K, Kalodimos, C.G. | | Deposit date: | 2018-02-21 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of chaperone-substrate complexes docked onto the export gate in a type III secretion system.
Nat Commun, 9, 2018
|
|
6DFY
 
 | | Remodeled crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*A)-3'), Double homeobox protein 4 | | Authors: | Aihara, H, Shi, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Comment on structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
1FUF
 
 | | CRYSTAL STRUCTURE OF A 14BP RNA OLIGONUCLEOTIDE CONTAINING DOUBLE UU BULGES: A NOVEL INTRAMOLECULAR U*(AU) BASE TRIPLE | | Descriptor: | (5'-R(*GP*GP*UP*AP*UP*UP*UP*CP*GP*GP*UP*AP*(CBR)P*C)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Deng, J, Xiong, Y, Sudarsanakumar, C, Shi, K, Sundaralingam, M. | | Deposit date: | 2000-09-15 | | Release date: | 2001-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two forms of a 14-mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A x U) base triple.
RNA, 7, 2001
|
|
7MC6
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MC5
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1IK5
 
 | | Crystal Structure of a 14mer RNA Containing Double UU Bulges in Two Crystal Forms: A Novel U*(AU) Intramolecular Base Triple | | Descriptor: | 5'-R(*GP*GP*UP*AP*UP*UP*UP*CP*GP*GP*UP*AP*(CBR)P*C)-3', 5'-R(*GP*GP*UP*AP*UP*UP*UP*UP*GP*GP*UP*AP*(CBR)P*C)-3', MAGNESIUM ION | | Authors: | Deng, J, Xiong, Y, Sudarsanakumar, C, Shi, K, Sundaralingam, M. | | Deposit date: | 2001-05-02 | | Release date: | 2001-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two forms of a 14-mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A x U) base triple.
RNA, 7, 2001
|
|
1J6S
 
 | | Crystal Structure of an RNA Tetraplex (UGAGGU)4 with A-tetrads, G-tetrads, U-tetrads and G-U octads | | Descriptor: | 5'-R(*(BRUP*GP*AP*GP*GP*U)-3', BARIUM ION, SODIUM ION | | Authors: | Pan, B, Xiong, Y, Shi, K, Deng, J, Sundaralingam, M. | | Deposit date: | 2002-07-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads:
implications for specific binding in RNA tetraplexes
Structure, 11, 2003
|
|
5WLY
 
 | | E. coli LpxH- 8 mutations | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The substrate-binding cap of the UDP-diacylglucosamine pyrophosphatase LpxH is highly flexible, enabling facile substrate binding and product release.
J. Biol. Chem., 293, 2018
|
|
7KM6
 
 | | APOBEC3B antibody 5G7 Fv-clasp | | Descriptor: | 1,2-ETHANEDIOL, 5G7 human monoclonal FAB heavy chain, 5G7 human monoclonal FAB light chain, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Characterization of a Minimal Antibody against Human APOBEC3B.
Viruses, 13, 2021
|
|
4PT6
 
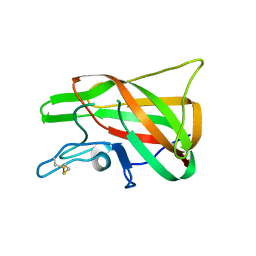 | | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities | | Descriptor: | Coagulation factor VIII | | Authors: | Kym, G, Shi, K, Kamajaya, A, Hamdouche, S, Kostecki, J.S, Wannier, T, Li, B, Nikolovski, P, Mayo, S.L. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities
To be Published
|
|
418D
 
 | |
2GRB
 
 | |
5HX5
 
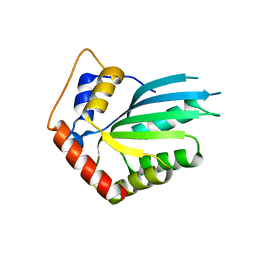 | | APOBEC3F Catalytic Domain Crystal Structure | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3F, ZINC ION | | Authors: | Shaban, N.M, Shi, K, Aihara, H, Harris, R.S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 1.92 Angstrom Zinc-Free APOBEC3F Catalytic Domain Crystal Structure.
J.Mol.Biol., 428, 2016
|
|
5HX4
 
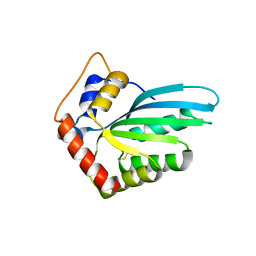 | |
3HL6
 
 | | Staphylococcus aureus pathogenicity island 3 ORF9 protein | | Descriptor: | CHLORIDE ION, Pathogenicity island protein | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional studies of a pathogenicity island protein from Staphylococcus aureus
To be Published
|
|
3K55
 
 | | Structure of beta hairpin deletion mutant of beta toxin from Staphylococcus aureus | | Descriptor: | Beta-hemolysin, CHLORIDE ION, SODIUM ION | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-10-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure of a mutant beta toxin from Staphylococcus aureus reveals domain swapping and conformational flexibility
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3LMX
 
 | | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Purpero, V.M, Lipscomb, J.D, Shi, K. | | Deposit date: | 2010-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published
|
|
5E96
 
 | | Crystal structure of aminoglycoside 6'-acetyltransferase type Ii | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside 6'-acetyltransferase, PHOSPHATE ION | | Authors: | Berghuis, A.M, Burk, D.L, Baettig, O.M, Shi, K. | | Deposit date: | 2015-10-14 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive characterization of ligand-induced plasticity changes in a dimeric enzyme.
Febs J., 283, 2016
|
|
5EJK
 
 | | Crystal structure of the Rous sarcoma virus intasome | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*T)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*CP*TP*C)-3'), ... | | Authors: | Yin, Z, Shi, K, Banerjee, S, Aihara, H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the Rous sarcoma virus intasome.
Nature, 530, 2016
|
|
8FR5
 
 | | Crystal structure of the Human Smacovirus 1 Rep domain | | Descriptor: | MANGANESE (II) ION, Rep, SODIUM ION | | Authors: | Limon, L.K, Shi, K, Dao, A, Rugloski, J, Tompkins, K.J, Aihara, H, Gordon, W.R, Evans IIII, R.L. | | Deposit date: | 2023-01-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The crystal structure of the human smacovirus 1 Rep domain.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
