4PRV
 
 | |
1BLE
 
 | |
1NRZ
 
 | |
1OAT
 
 | | ORNITHINE AMINOTRANSFERASE | | Descriptor: | ORNITHINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shen, B.W, Schirmer, T, Jansonius, J.N. | | Deposit date: | 1997-03-26 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human recombinant ornithine aminotransferase.
J.Mol.Biol., 277, 1998
|
|
1OXP
 
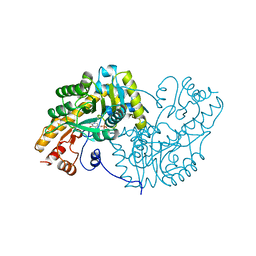 | | ASPARTATE AMINOTRANSFERASE, H-ASP COMPLEX, CLOSED CONFORMATION | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Hohenester, E, Schirmer, T, Jansonius, J.N. | | Deposit date: | 1995-12-23 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and solution studies of oxime adducts of mitochondrial aspartate aminotransferase.
Eur.J.Biochem., 236, 1996
|
|
1OXO
 
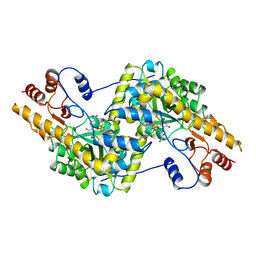 | | ASPARTATE AMINOTRANSFERASE, H-ASP COMPLEX, OPEN CONFORMATION | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Hohenester, E, Schirmer, T, Jansonius, J.N. | | Deposit date: | 1995-12-23 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and solution studies of oxime adducts of mitochondrial aspartate aminotransferase.
Eur.J.Biochem., 236, 1996
|
|
1OSM
 
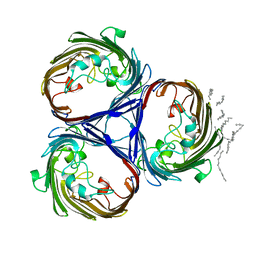 | | OSMOPORIN (OMPK36) FROM KLEBSIELLA PNEUMONIAE | | Descriptor: | DODECANE, OMPK36 | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1999-01-08 | | Release date: | 1999-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and functional characterization of OmpK36, the osmoporin of Klebsiella pneumoniae.
Structure Fold.Des., 7, 1999
|
|
1PDO
 
 | |
2BYL
 
 | | Structure of ornithine aminotransferase triple mutant Y85I Y55A G320F | | Descriptor: | ORNITHINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Markova, M, Peneff, C, Hewlins, M.J.E, Schirmer, T, John, R.A. | | Deposit date: | 2005-08-03 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Determinants of Substrate Specificity in Omega-Aminotransferases.
J.Biol.Chem., 280, 2005
|
|
2BYJ
 
 | | Ornithine aminotransferase mutant Y85I | | Descriptor: | ORNITHINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Markova, M, Peneff, C, Hewlins, M.J.E, Schirmer, T, John, R.A. | | Deposit date: | 2005-08-02 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Determinants of Substrate Specificity in Omega-Aminotransferases.
J.Biol.Chem., 280, 2005
|
|
1FCQ
 
 | | CRYSTAL STRUCTURE (MONOCLINIC) OF BEE VENOM HYALURONIDASE | | Descriptor: | HYALURONOGLUCOSAMINIDASE | | Authors: | Markovic-Housley, Z, Miglierini, G, Soldatova, L, Mueller, U, Schirmer, T. | | Deposit date: | 2000-07-19 | | Release date: | 2001-10-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of hyaluronidase, a major allergen of bee venom.
Structure Fold.Des., 8, 2000
|
|
1FCV
 
 | | CRYSTAL STRUCTURE OF BEE VENOM HYALURONIDASE IN COMPLEX WITH HYALURONIC ACID TETRAMER | | Descriptor: | HYALURONOGLUCOSAMINIDASE, alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid | | Authors: | Markovic-Housley, Z, Miglierini, G, Soldatova, L, Rizkallah, P.J, Mueller, U, Schirmer, T. | | Deposit date: | 2000-07-19 | | Release date: | 2001-10-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of hyaluronidase, a major allergen of bee venom.
Structure Fold.Des., 8, 2000
|
|
1FCU
 
 | | CRYSTAL STRUCTURE (TRIGONAL) OF BEE VENOM HYALURONIDASE | | Descriptor: | HYALURONOGLUCOSAMINIDASE | | Authors: | Markovic-Housley, Z, Miglierini, G, Soldatova, L, Rizkallah, P.J, Mueller, U, Schirmer, T. | | Deposit date: | 2000-07-19 | | Release date: | 2001-10-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of hyaluronidase, a major allergen of bee venom.
Structure Fold.Des., 8, 2000
|
|
2C32
 
 | |
3S6A
 
 | |
3SHG
 
 | |
3SE5
 
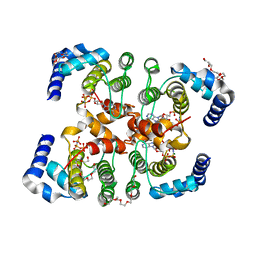 | | Fic protein from NEISSERIA MENINGITIDIS mutant delta8 in complex with AMPPNP | | Descriptor: | Cell filamentation protein Fic-related protein, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Goepfert, A, Stanger, F, Schirmer, T. | | Deposit date: | 2011-06-10 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins.
Nature, 482, 2012
|
|
3S83
 
 | | EAL domain of phosphodiesterase PdeA | | Descriptor: | GGDEF family protein, POTASSIUM ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of EAL domain from Caulobacter crescentus CB15
To be Published
|
|
3SN9
 
 | |
3T9O
 
 | | Regulatory CZB domain of DgcZ | | Descriptor: | Diguanylate cyclase DgcZ, ZINC ION | | Authors: | Zaehringer, F, Schirmer, T. | | Deposit date: | 2011-08-03 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and signaling mechanism of a zinc-sensory diguanylate cyclase.
Structure, 21, 2013
|
|
3TVK
 
 | | Diguanylate cyclase domain of DgcZ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), D(-)-TARTARIC ACID, Diguanylate cyclase DgcZ, ... | | Authors: | Zaehringer, F, Schirmer, T. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and signaling mechanism of a zinc-sensory diguanylate cyclase.
Structure, 21, 2013
|
|
3U2E
 
 | | EAL domain of phosphodiesterase PdeA in complex with 5'-pGpG and Mg++ | | Descriptor: | GGDEF family protein, MAGNESIUM ION, RNA (5'-R(P*GP*G)-3') | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-03 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | EAL domain from Caulobacter crescentus in complex with 5'-pGpG and Mg++
To be Published
|
|
2V0N
 
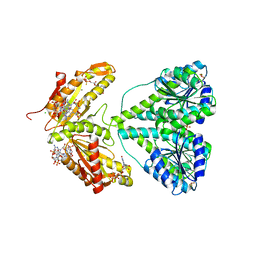 | | ACTIVATED RESPONSE REGULATOR PLED IN COMPLEX WITH C-DIGMP AND GTP- ALPHA-S | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, ... | | Authors: | Wassmann, P, Schirmer, T. | | Deposit date: | 2007-05-15 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of Bef3--Modified Response Regulator Pled: Implications for Diguanylate Cyclase Activation, Catalysis, and Feedback Inhibition
Structure, 15, 2007
|
|
2VY3
 
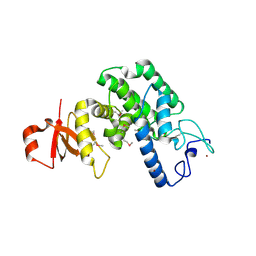 | | Type IV secretion system effector protein BepA | | Descriptor: | CELL FILAMENTATION PROTEIN, CHLORIDE ION, NICKEL (II) ION | | Authors: | Palanivelu, D.V, Schirmer, T. | | Deposit date: | 2008-07-17 | | Release date: | 2009-11-17 | | Last modified: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fic Domain Catalyzed Adenylylation: Insight Provided by the Structural Analysis of the Type Iv Secretion System Effector Bepa.
Protein Sci., 20, 2011
|
|
2VZ4
 
 | |
