6TNE
 
 | |
2OAT
 
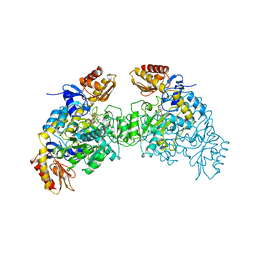 | | ORNITHINE AMINOTRANSFERASE COMPLEXED WITH 5-FLUOROMETHYLORNITHINE | | Descriptor: | 1-AMINO-7-(2-METHYL-3-OXIDO-5-((PHOSPHONOXY)METHYL)-4-PYRIDOXAL-5-OXO-6-HEPTENATE, ORNITHINE AMINOTRANSFERASE | | Authors: | Storici, P, Schirmer, T. | | Deposit date: | 1998-05-07 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human ornithine aminotransferase complexed with the highly specific and potent inhibitor 5-fluoromethylornithine.
J.Mol.Biol., 285, 1999
|
|
3ZP9
 
 | |
4XJY
 
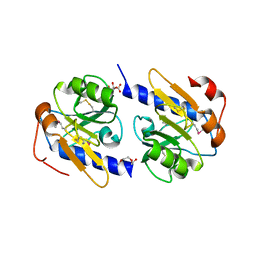 | | Periplasmic repressor protein YfiR | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, L(+)-TARTARIC ACID, YfiR | | Authors: | Kauer, S, Schirmer, T. | | Deposit date: | 2015-01-09 | | Release date: | 2016-01-20 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Periplasmic repressor protein YfiR at 1.8 Angstroms resolution
To Be Published
|
|
5IDM
 
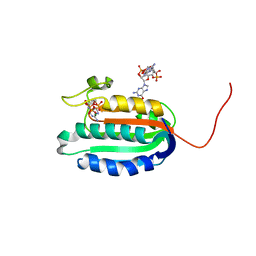 | | Bifunctional histidine kinase CckA (domain, CA) in complex with c-di-GMP and AMPPNP/Mg2+ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Cell cycle histidine kinase CckA, MAGNESIUM ION, ... | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2022-09-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking.
Sci Adv, 2, 2016
|
|
1OPF
 
 | |
5JFF
 
 | | E. coli EcFicT mutant G55R in complex with EcFicA | | Descriptor: | CHLORIDE ION, Probable adenosine monophosphate-protein transferase fic, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Stanger, F.V, Schirmer, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Escherichia coli Fic Toxin-Like Protein in Complex with Its Cognate Antitoxin.
Plos One, 11, 2016
|
|
5JFZ
 
 | | E. coli EcFicT in complex with EcFicA mutant E28G | | Descriptor: | Probable adenosine monophosphate-protein transferase fic, Uncharacterized protein YhfG | | Authors: | Stanger, F.V, Schirmer, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Escherichia coli Fic Toxin-Like Protein in Complex with Its Cognate Antitoxin.
Plos One, 11, 2016
|
|
1GFM
 
 | | OMPF PORIN (MUTANT D113G) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
1GFP
 
 | | OMPF PORIN (MUTANT R42C) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
1GFN
 
 | | OMPF PORIN DELETION (MUTANT DELTA 109-114) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
1GFO
 
 | | OMPF PORIN (MUTANT R132P) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
1GFQ
 
 | | OMPF PORIN (MUTANT R82C) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
1MPQ
 
 | | MALTOPORIN TREHALOSE COMPLEX | | Descriptor: | MALTOPORIN, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1997-03-24 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin.
J.Mol.Biol., 272, 1997
|
|
6QKI
 
 | | Native structure of EgtB from Chloracidobacterium thermophilum, a type II sulfoxide synthase | | Descriptor: | FE (III) ION, Uncharacterized protein | | Authors: | Stampfli, A.R, Badri, B.N, Schirmer, T, Seebeck, F.P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-27 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | An Alternative Active Site Architecture for O2Activation in the Ergothioneine Biosynthetic EgtB from Chloracidobacterium thermophilum.
J.Am.Chem.Soc., 141, 2019
|
|
6QKJ
 
 | | EgtB from Chloracidobacterium thermophilum, a type II sulfoxide synthase in complex with N,N,N-trimethyl-histidine | | Descriptor: | CHLORIDE ION, FE (III) ION, IMIDAZOLE, ... | | Authors: | Stampfli, A.R, Badri, B.N, Schirmer, T, Seebeck, F.P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-27 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Alternative Active Site Architecture for O2Activation in the Ergothioneine Biosynthetic EgtB from Chloracidobacterium thermophilum.
J.Am.Chem.Soc., 141, 2019
|
|
3ZC7
 
 | |
3ZCN
 
 | |
3ZCB
 
 | |
6QRJ
 
 | |
3ZLM
 
 | |
3ZEC
 
 | |
6QRL
 
 | | Crystal structure of ShkA _Rec1 in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Hybrid kinase, SULFATE ION | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2019-02-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hybrid histidine kinase activation by cyclic di-GMP-mediated domain liberation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4GJV
 
 | | Streptavidin-S112H | | Descriptor: | CHLORIDE ION, Rhodium, Streptavidin, ... | | Authors: | Heinisch, T, Schirmer, T. | | Deposit date: | 2012-08-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dual anchoring strategy for the localization and activation of artificial metalloenzymes based on the biotin-streptavidin technology.
J.Am.Chem.Soc., 135, 2013
|
|
4GJS
 
 | | Streptavidin-K121H | | Descriptor: | Rhodium, Streptavidin, trichloro{(1,2,3,4,5-eta)-1,2,3,4-tetramethyl-5-[2-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)ethyl]cyclopentadienyl}rhodium(1+) | | Authors: | Heinisch, T, Schirmer, T. | | Deposit date: | 2012-08-10 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A dual anchoring strategy for the localization and activation of artificial metalloenzymes based on the biotin-streptavidin technology.
J.Am.Chem.Soc., 135, 2013
|
|
