6EKZ
 
 | |
6EKW
 
 | |
6EKX
 
 | |
6EKY
 
 | |
6EL4
 
 | |
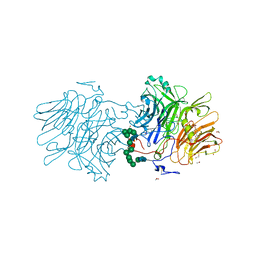
6FJG
 
 | |
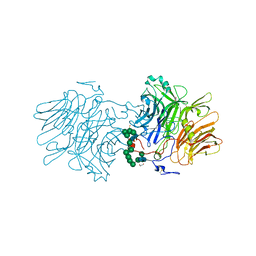
6FJE
 
 | |
6S6Z
 
 | | Structure of beta-Galactosidase from Thermotoga maritima | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Miguez-Amil, S, Jimenez-Ortega, E, Ramirez Escudero, M, Sanz-Aparicio, J, Fernandez-Leiro, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | The cryo-EM Structure ofThermotoga maritimabeta-Galactosidase: Quaternary Structure Guides Protein Engineering.
Acs Chem.Biol., 15, 2020
|
|
6SD0
 
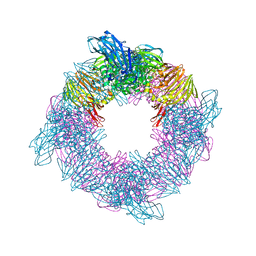 | |
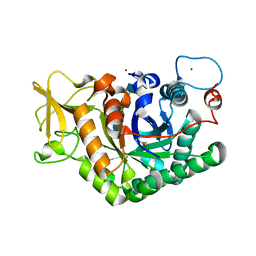
7AKQ
 
 | |
1QGZ
 
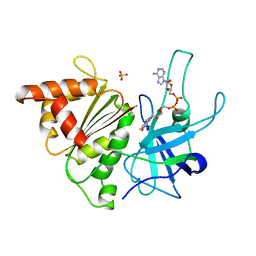 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 78 REPLACED BY ASP (L78D) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
1QH0
 
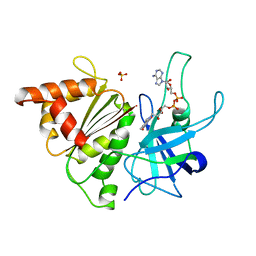 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 76 MUTATED BY ASP AND LEU 78 MUTATED BY ASP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
1QGY
 
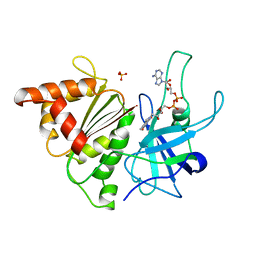 | | Ferredoxin:NADP+ reductase mutant with Lys 75 replaced by Glu (K75E) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP+ reductase, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of interactions for complex formation between Ferredoxin-NADP+ reductase and its protein partners
Proteins, 59, 2005
|
|
6QWI
 
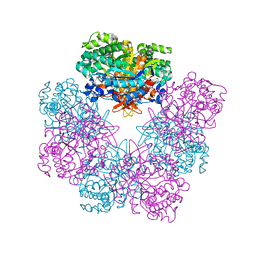 | | Structure of beta-glucosidase A from Paenibacillus polymyxa complexed with multivalent inhibitors. | | Descriptor: | (2~{S},3~{S},4~{R})-2-[[4-[4-(2-ethoxyethoxy)phenyl]-1,2,3-triazol-1-yl]methyl]pyrrolidine-3,4-diol, Beta-glucosidase A | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of the inhibition of GH1 beta-glucosidases by multivalent pyrrolidine iminosugars.
Bioorg.Chem., 89, 2019
|
|
6R4K
 
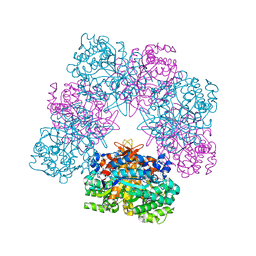 | | Structure of beta-glucosidase A from Paenibacillus polymyxa complexed with a monovalent inhibitor | | Descriptor: | (2~{S},3~{S},4~{R})-2-[[4-[4-[2-[2-(2-azanylidenehydrazinyl)ethoxy]ethoxy]phenyl]-1,2,3-triazol-1-yl]methyl]pyrrolidine-3,4-diol, Beta-glucosidase A | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of the inhibition of GH1 beta-glucosidases by multivalent pyrrolidine iminosugars.
Bioorg.Chem., 89, 2019
|
|
6RB0
 
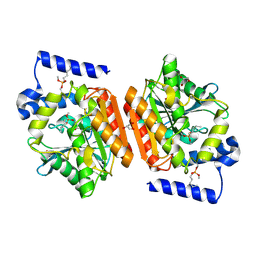 | |
6RKY
 
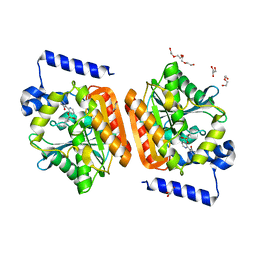 | |
6SUD
 
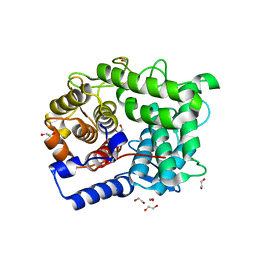 | | Structure of L320A mutant of Rex8A from Paenibacillus barcinonensis complexed with xylose. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Reducing-end xylose-releasing exo-oligoxylanase Rex8A, ... | | Authors: | Jimenez-Ortega, E, Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-09-13 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of the reducing-end xylose-releasing exo-oligoxylanase Rex8A from Paenibacillus barcinonensis BP-23 deciphers its molecular specificity.
Febs J., 287, 2020
|
|
6SHY
 
 | |
6SRD
 
 | |
6TO0
 
 | |
6TPP
 
 | |
6TOW
 
 | |
6TRH
 
 | |
6S82
 
 | | Structure Of D80A-Fructofuranosidase From Xanthophyllomyces Dendrorhous Complexed With Tris-buffer molecule And hydroquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deciphering the molecular specificity of phenolic compounds as inhibitors or glycosyl acceptors of beta-fructofuranosidase from Xanthophyllomyces dendrorhous.
Sci Rep, 9, 2019
|
|
