5SCM
 
 | |
5SCN
 
 | |
5SD1
 
 | |
5SCS
 
 | |
5SCX
 
 | |
5SCV
 
 | |
5SD8
 
 | |
5SCQ
 
 | |
5SD5
 
 | |
5SD2
 
 | |
5SCY
 
 | |
5SCU
 
 | |
5SD6
 
 | |
5T8S
 
 | | Crystal Structure of a S-adenosylmethionine Synthase from Neisseria gonorrhoeae with bound S-adenosylmethionine, AMP, Pyrophosphate, Phosphate, and Magnesium | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2016-09-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a S-adenosylmethionine Synthase from Neisseria gonorrhoeae with bound S-adenosylmethionine, AMP, Pyrophosphate, Phosphate, and Magnesium
to be published
|
|
5T5Q
 
 | |
5T8T
 
 | |
4TM5
 
 | |
4TVI
 
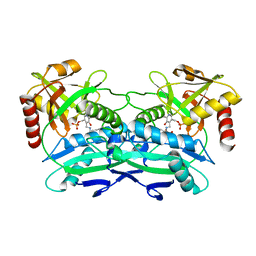 | |
4TWR
 
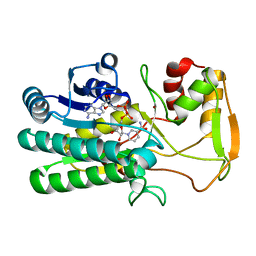 | | Structure of UDP-glucose 4-epimerase from Brucella abortus | | Descriptor: | NAD binding site:NAD-dependent epimerase/dehydratase:UDP-glucose 4-epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-07-01 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of UDP-glucose 4-epimerase from Brucella melitensis
To Be Published
|
|
2KWL
 
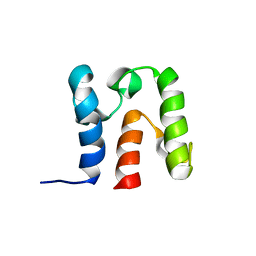 | | Solution Structure of acyl carrier protein from Borrelia burgdorferi | | Descriptor: | Acyl carrier protein | | Authors: | Barnwal, R, Vittal, V, Moody, J, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-04-12 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of an acyl-carrier protein from Borrelia burgdorferi.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2MJ3
 
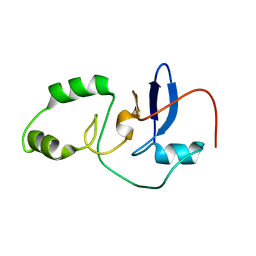 | |
2LXF
 
 | |
2M4Y
 
 | |
2M0N
 
 | |
2MRL
 
 | |
