4R85
 
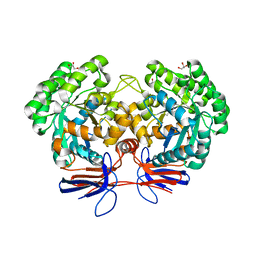 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine | | 分子名称: | 5-methylcytosine, Cytosine deaminase, FE (II) ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | 登録日 | 2014-08-29 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine
To be Published
|
|
4R7W
 
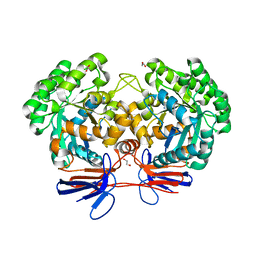 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine | | 分子名称: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, 1,2-ETHANEDIOL, Cytosine deaminase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | 登録日 | 2014-08-28 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine
To be Published
|
|
4RDW
 
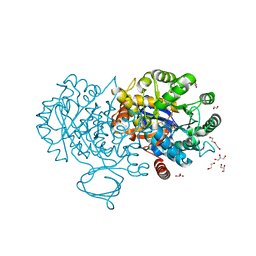 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | 登録日 | 2014-09-19 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.591 Å) | | 主引用文献 | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid
To be Published
|
|
4M51
 
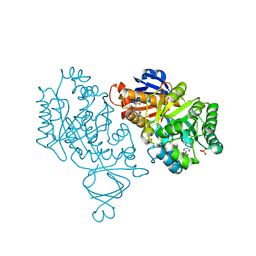 | | Crystal structure of amidohydrolase nis_0429 (ser145ala mutant) from nitratiruptor sp. sb155-2 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase family protein, BENZOIC ACID, ... | | 著者 | Patskovsky, Y, Toro, R, Gobble, A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-08-07 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes.
Biochemistry, 52, 2013
|
|
4RZB
 
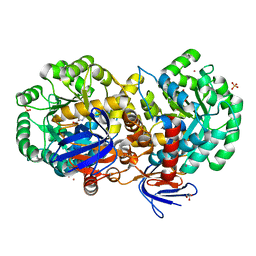 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate, SOAKED WITH MERCURY | | 分子名称: | GLYCEROL, MERCURY (II) ION, N-[(E)-iminomethyl]-L-aspartic acid, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | 登録日 | 2014-12-19 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.863 Å) | | 主引用文献 | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
1JDB
 
 | | CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIA COLI | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE, CHLORIDE ION, ... | | 著者 | Thoden, J.B, Holden, H.M, Wesenberg, G, Raushel, F.M, Rayment, I. | | 登録日 | 1997-03-25 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The structure of carbamoyl phosphate synthetase determined to 2.1 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3ID7
 
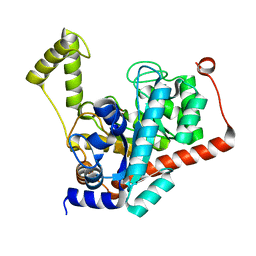 | | Crystal structure of renal dipeptidase from Streptomyces coelicolor A3(2) | | 分子名称: | CHLORIDE ION, Dipeptidase, ZINC ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | 登録日 | 2009-07-20 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3IJ6
 
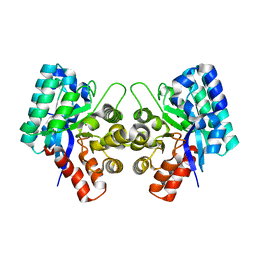 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidophilus | | 分子名称: | SODIUM ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE, ZINC ION | | 著者 | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-08-03 | | 公開日 | 2009-08-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidopphilus
To be Published
|
|
3ICJ
 
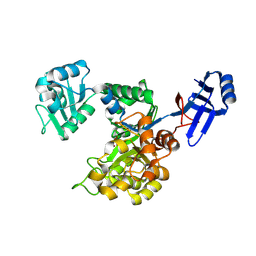 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus furiosus | | 分子名称: | ZINC ION, uncharacterized metal-dependent hydrolase | | 著者 | Bonanno, J.B, Patskovsky, Y, Freeman, J, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-17 | | 公開日 | 2009-07-28 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Furiosus
To be Published
|
|
3IGH
 
 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus horikoshii ot3 | | 分子名称: | SULFATE ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE | | 著者 | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-27 | | 公開日 | 2009-08-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Horikoshii
To be Published
|
|
3IRS
 
 | | CRYSTAL STRUCTURE OF UNCHARACTERIZED TIM-BARREL PROTEIN BB4693 FROM Bordetella bronchiseptica | | 分子名称: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | 著者 | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Do, J, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-08-24 | | 公開日 | 2009-09-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF UNCHARACTERIZED HYDROLASE FROM Bordetella bronchiseptica
To be Published
|
|
3ITC
 
 | | Crystal structure of Sco3058 with bound citrate and glycerol | | 分子名称: | CITRIC ACID, GLYCEROL, ZINC ION, ... | | 著者 | Nguyen, T.T, Cummings, J.A, Tsai, C.-L, Barondeau, D.P, Raushel, F.M. | | 登録日 | 2009-08-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase
Biochemistry, 49, 2010
|
|
3K5X
 
 | | Crystal structure of dipeptidase from Streptomics coelicolor complexed with phosphinate pseudodipeptide L-Ala-D-Asp at 1.4A resolution. | | 分子名称: | Dipeptidase, ZINC ION, phosphinate pseudodipeptide L-Ala-D-Asp | | 著者 | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | 登録日 | 2009-10-08 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3LY0
 
 | | Crystal structure of metallo peptidase from Rhodobacter sphaeroides liganded with phosphinate mimic of dipeptide L-Ala-D-Ala | | 分子名称: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Dipeptidase AC. Metallo peptidase. MEROPS family M19, ZINC ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-02-25 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | Crystal structure of metallo peptidase from Rhodobacter sphaeroides
liganded with phosphinate mimic of dipeptide L-Ala-D-Ala
To be Published
|
|
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | 分子名称: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | 著者 | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-04-15 | | 公開日 | 2010-04-28 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3MDW
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | 分子名称: | GLYCEROL, N-[(E)-iminomethyl]-L-aspartic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | 登録日 | 2010-03-30 | | 公開日 | 2011-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8979 Å) | | 主引用文献 | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
3MDU
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutamate | | 分子名称: | GLYCEROL, N-carbamimidoyl-L-glutamic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | 登録日 | 2010-03-30 | | 公開日 | 2011-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4003 Å) | | 主引用文献 | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
3MTW
 
 | | Crystal structure of L-Lysine, L-Arginine carboxypeptidase Cc2672 from Caulobacter Crescentus CB15 complexed with N-methyl phosphonate derivative of L-Arginine | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L-Arginine carboxypeptidase Cc2672, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | 登録日 | 2010-05-01 | | 公開日 | 2010-07-28 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Functional Identification and Structure Determination of Two Novel Prolidases from cog1228 in the Amidohydrolase Superfamily
Biochemistry, 49, 2010
|
|
3N2C
 
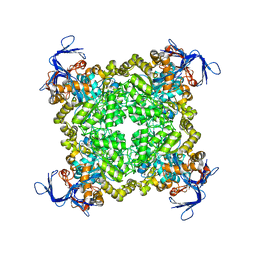 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | 分子名称: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | 著者 | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-05-17 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3NEH
 
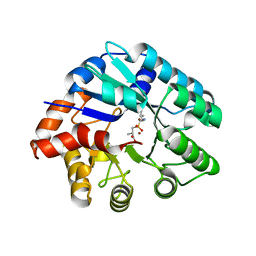 | | Crystal structure of the protein LMO2462 from Listeria monocytogenes complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala | | 分子名称: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Renal dipeptidase family protein, ZINC ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-06-08 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.642 Å) | | 主引用文献 | Crystal structure of the protein LMO2462 from Listeria monocytogenes
complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala
To be Published
|
|
4DO7
 
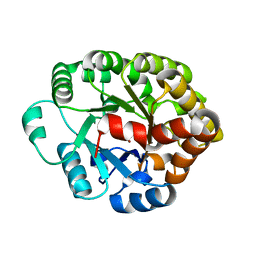 | | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2 | | 分子名称: | Amidohydrolase 2, SULFATE ION, ZINC ION | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-09 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2
to be published
|
|
4DI9
 
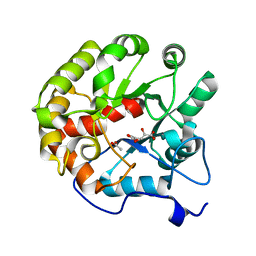 | | CRYSTAL STRUCTURE OF THE D248A mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 6.5 | | 分子名称: | (1E,3Z)-4-hydroxybuta-1,3-diene-1,2,4-tricarboxylic acid, 2-pyrone-4,6-dicarbaxylate hydrolase, ACETATE ION | | 著者 | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | 登録日 | 2012-01-11 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4DIA
 
 | | CRYSTAL STRUCTURE OF THE D248N mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 4.6 | | 分子名称: | 2-pyrone-4,6-dicarbaxylate hydrolase | | 著者 | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | 登録日 | 2012-01-11 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4DLM
 
 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121 | | 分子名称: | Amidohydrolase 2, ZINC ION | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-06 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121
to be published
|
|
4DLF
 
 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P3221 | | 分子名称: | Amidohydrolase 2, GLYCEROL, UNKNOWN ATOM OR ION, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-06 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (target efi-500235) with bound ZN, space group P3221
to be published
|
|
