6H6E
 
 | | PTC3 holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3) | | Descriptor: | TcdA1, TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
6RW8
 
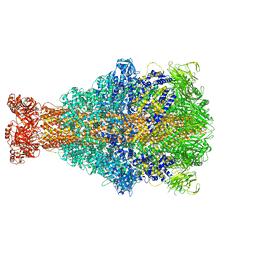 | | Cryo-EM structure of Xenorhabdus nematophila XptA1 | | Descriptor: | A component of insecticidal toxin complex (Tc) | | Authors: | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
6RW9
 
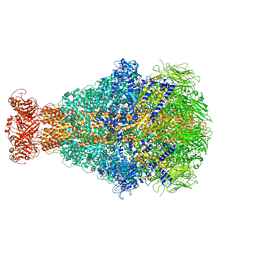 | | Cryo-EM structure of Morganella morganii TcdA4 | | Descriptor: | Insecticidal toxin protein TcdA4 | | Authors: | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
6RWB
 
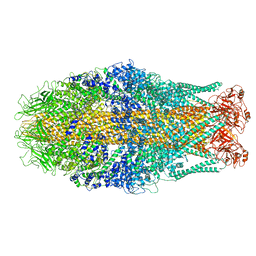 | | Cryo-EM structure of Yersinia pseudotuberculosis TcaA-TcaB | | Descriptor: | Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit,Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit,Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit,Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit | | Authors: | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
6RWA
 
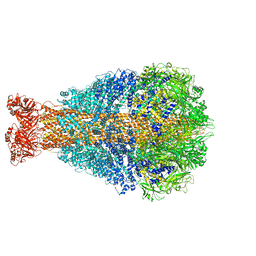 | | Cryo-EM structure of Photorhabdus luminescens TcdA4 | | Descriptor: | TcdA4 | | Authors: | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
6RW6
 
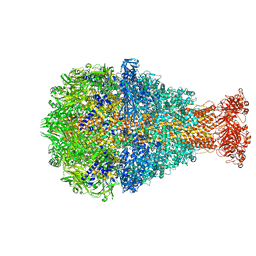 | | Cryo-EM structure of Photorhabdus luminescens TcdA1 | | Descriptor: | TcdA1 | | Authors: | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
6SFW
 
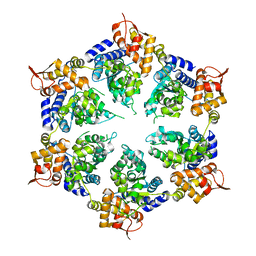 | |
6SFX
 
 | |
6SUP
 
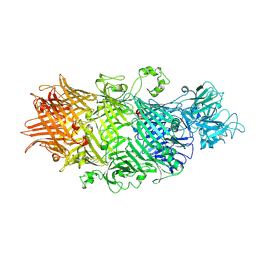 | | Crystal Structure of TcdB2-TccC3-Cdc42 | | Descriptor: | MAGNESIUM ION, TcdB2,TccC3,Cell division control protein 42 homolog | | Authors: | Roderer, D, Schubert, E, Sitsel, O, Raunser, S. | | Deposit date: | 2019-09-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Towards the application of Tc toxins as a universal protein translocation system.
Nat Commun, 10, 2019
|
|
6SUQ
 
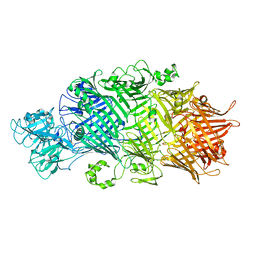 | | Crystal Structure of TcdB2-TccC3-TEV | | Descriptor: | TcdB2,TccC3,Genome polyprotein | | Authors: | Roderer, D, Schubert, E, Sitsel, O, Raunser, S. | | Deposit date: | 2019-09-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Towards the application of Tc toxins as a universal protein translocation system.
Nat Commun, 10, 2019
|
|
6SUF
 
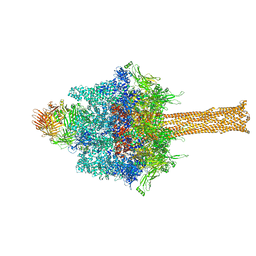 | |
7PXH
 
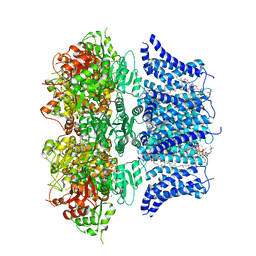 | | Emodepside-bound Drosophila Slo channel | | Descriptor: | (3~{S},6~{R},9~{S},12~{R},15~{S},18~{R},21~{S},24~{R})-4,6,10,16,18,22-hexamethyl-3,9,15,21-tetrakis(2-methylpropyl)-12,24-bis[(4-morpholin-4-ylphenyl)methyl]-1,7,13,19-tetraoxa-4,10,16,22-tetrazacyclotetracosane-2,5,8,11,14,17,20,23-octone, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CALCIUM ION, ... | | Authors: | Raisch, T, Brockmann, A, Ebbinghaus-Kintscher, U, Freigang, J, Gutbrod, O, Kubicek, J, Maertens, B, Hofnagel, O, Raunser, S. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Small molecule modulation of the Drosophila Slo channel elucidated by cryo-EM.
Nat Commun, 12, 2021
|
|
7PXG
 
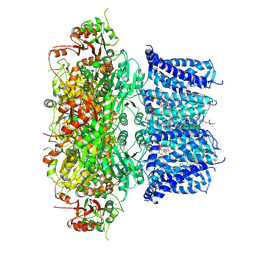 | | Verruculogen-bound Drosophila Slo channel | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Raisch, T, Brockmann, A, Ebbinghaus-Kintscher, U, Freigang, J, Gutbrod, O, Kubicek, J, Maertens, B, Hofnagel, O, Raunser, S. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Small molecule modulation of the Drosophila Slo channel elucidated by cryo-EM.
Nat Commun, 12, 2021
|
|
7PXF
 
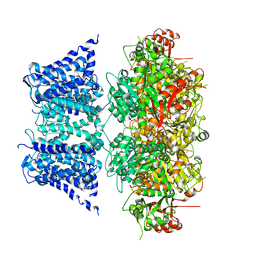 | | Ca2+ free Drosophila Slo channel | | Descriptor: | Isoform J of Calcium-activated potassium channel slowpoke, MAGNESIUM ION, POTASSIUM ION | | Authors: | Raisch, T, Brockmann, A, Ebbinghaus-Kintscher, U, Freigang, J, Gutbrod, O, Kubicek, J, Maertens, B, Hofnagel, O, Raunser, S. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Small molecule modulation of the Drosophila Slo channel elucidated by cryo-EM.
Nat Commun, 12, 2021
|
|
7PXE
 
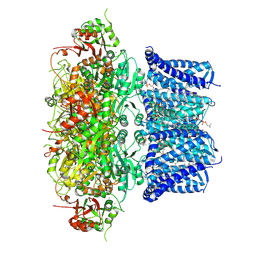 | | Ca2+ bound Drosophila Slo channel | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Raisch, T, Brockmann, A, Ebbinghaus-Kintscher, U, Freigang, J, Gutbrod, O, Kubicek, J, Maertens, B, Hofnagel, O, Raunser, S. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Small molecule modulation of the Drosophila Slo channel elucidated by cryo-EM.
Nat Commun, 12, 2021
|
|
7PLZ
 
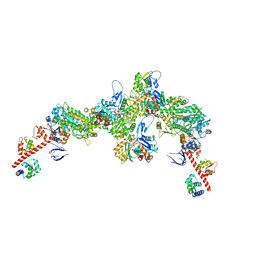 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 3er/2er, young JASP-stabilized F-actin) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PMD
 
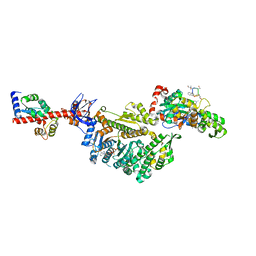 | | Cryo-EM structure of the actomyosin-V complex in the post-rigor transition state (AppNHp, central 1er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PLT
 
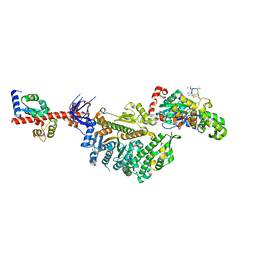 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PM3
 
 | | Cryo-EM structure of young JASP-stabilized F-actin (central 3er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PMF
 
 | | Cryo-EM structure of the actomyosin-V complex in the post-rigor transition state (AppNHp, central 1er, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PMA
 
 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er, class 5) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PML
 
 | | Cryo-EM structure of the actomyosin-V complex in the post-rigor transition state (AppNHp, central 1er, class 8) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PM9
 
 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er, class 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PLU
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 3er/2er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PMH
 
 | | Cryo-EM structure of the actomyosin-V complex in the post-rigor transition state (AppNHp, central 1er, class 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
