7O9U
 
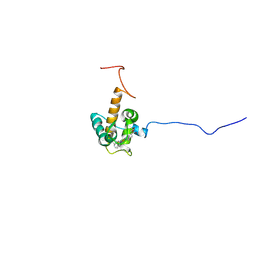 | | Solution structure of oxidized cytochrome c552 from Thioalkalivibrio paradoxus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Britikov, V.V, Britikova, E.V, Altukhov, D.A, Timofeev, V.I, Dergousova, N.I, Rakitina, T.V, Tikhonova, T.V, Usanov, S.A, Popov, V.O, Bocharov, E.V. | | Deposit date: | 2021-04-17 | | Release date: | 2021-05-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Unusual Cytochrome c 552 from Thioalkalivibrio paradoxus : Solution NMR Structure and Interaction with Thiocyanate Dehydrogenase.
Int J Mol Sci, 23, 2022
|
|
3OWM
 
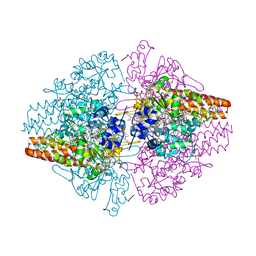 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with hydroxylamine | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-09-20 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding of sulfite by the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase
To be Published
|
|
3RKH
 
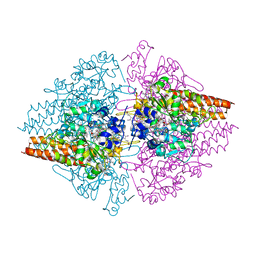 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite (full occupancy) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RR5
 
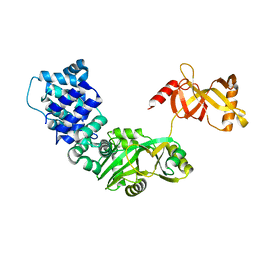 | | DNA ligase from the archaeon Thermococcus sp. 1519 | | Descriptor: | DNA ligase, MAGNESIUM ION | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Popov, V.O, Polyakov, K.M, Ravin, N.V, Shabalin, I.G, Skryabin, K.G, Stekhanova, T.N, Kovalchuk, M.V. | | Deposit date: | 2011-04-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.018 Å) | | Cite: | ATP-dependent DNA ligase from Thermococcus sp. 1519 displays a new arrangement of the OB-fold domain.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3S7W
 
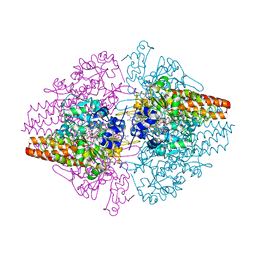 | | Structure of the TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with an oxidized Gln360 in a complex with hydroxylamine | | Descriptor: | AZIDE ION, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2011-05-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with an oxidized Gln360 in a complex with hydroxylamine
To be Published
|
|
3SCE
 
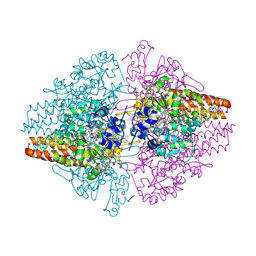 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with a covalent bond between the CE1 atom of Tyr303 and the CG atom of Gln360 (TvNiRb) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5CE8
 
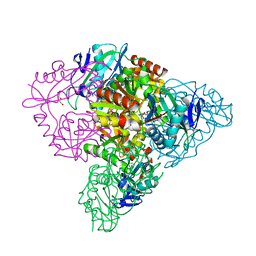 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Thermoproteus uzoniensis | | Descriptor: | Branched-chain amino acid aminotransferase, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-07-06 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First structure of archaeal branched-chain amino acid aminotransferase from Thermoproteus uzoniensis specific for L-amino acids and R-amines.
Extremophiles, 20, 2016
|
|
5CM0
 
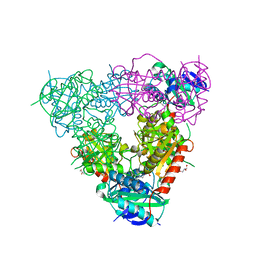 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Geoglobus acetivorans | | Descriptor: | Branched-chain transaminase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-07-16 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
5E25
 
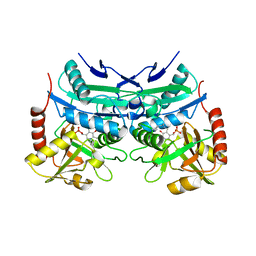 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Geoglobus acetivorans complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, PYRIDOXAL-5'-PHOSPHATE, branched-chain aminotransferase | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
5EEB
 
 | | Apo form of thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 | | Descriptor: | Aldehyde dehydrogenase | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-10-22 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.038 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
8YQ4
 
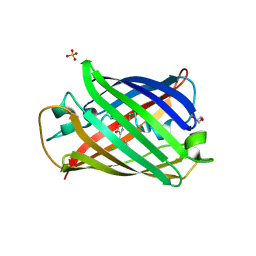 | | Structure of mBaoJin2 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Minyaev, M.E, Kuzmicheva, T.P, Vlaskina, A.V, Popov, V.O, Pyatkevich, K.D, Subach, F.V. | | Deposit date: | 2024-03-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mBaoJin2
To Be Published
|
|
4EQ5
 
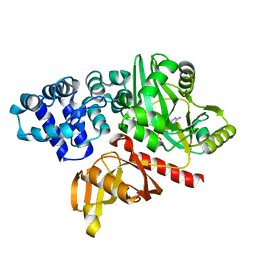 | | DNA ligase from the archaeon Thermococcus sibiricus | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase | | Authors: | Petrova, T, Bezsudnova, E.Y, Dorokhov, B.D, Slutskaya, E.S, Polyakov, K.M, Dorovatovskiy, P.V, Ravin, N.V, Skryabin, K.G, Kovalchuk, M.V, Popov, V.O. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Expression, purification, crystallization and preliminary crystallographic analysis of a thermostable DNA ligase from the archaeon Thermococcus sibiricus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4FFK
 
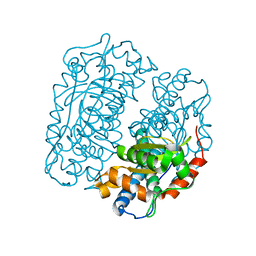 | | X-ray structure of iron superoxide dismutase from Acidilobus saccharovorans | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Safonova, T.N, Slutskaya, E.S, Dorovatovsky, P.V, Bezsudnova, E.Yu, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O, Polyakov, K.M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray structure of iron superoxide dismutase from Acidilobus saccharovorans
TO BE PUBLISHED
|
|
4FKC
 
 | | Recombinant prolidase from Thermococcus sibiricus | | Descriptor: | CADMIUM ION, Xaa-Pro aminopeptidase | | Authors: | Trofimov, A.A, Slutskaya, E.S, Polyakov, K.M, Dorovatovskii, P.V, Gumerov, V.M, Popov, V.O. | | Deposit date: | 2012-06-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Influence of intermolecular contacts on the structure of recombinant prolidase from Thermococcus sibiricus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4H73
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICKEL (II) ION | | Authors: | Petrova, T, Shabalin, I.G, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. complexed with NADP+
To be Published
|
|
4HA4
 
 | | Structure of beta-glycosidase from Acidilobus saccharovorans in complex with glycerol | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Beta-galactosidase, GLYCEROL, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonov, A.V, Bezsudnova, E.Y, Dorovatovskii, P.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-10 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structures of beta-glycosidase from Acidilobus saccharovorans in complexes with tris and glycerol.
Dokl.Biochem.Biophys., 449
|
|
4HA3
 
 | | Structure of beta-glycosidase from Acidilobus saccharovorans in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonov, A.V, Bezsudnova, E.Y, Dorovatovskii, P.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structures of beta-glycosidase from Acidilobus saccharovorans in complexes with tris and glycerol.
Dokl.Biochem.Biophys., 449
|
|
4H05
 
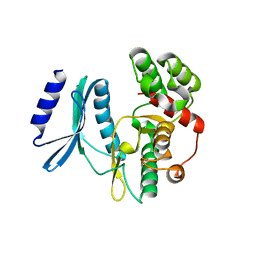 | | Crystal structure of aminoglycoside-3'-phosphotransferase of type VIII | | Descriptor: | Aminoglycoside-O-phosphotransferase VIII | | Authors: | Boyko, K.M, Gorbacheva, M.A, Danilenko, V.N, Alekseeva, M.G, Korzhenevskiy, D.A, Dorovatovskiy, P.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2012-09-07 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of the novel aminoglycoside phosphotransferase AphVIII from Streptomyces rimosus with enzymatic activity modulated by phosphorylation.
Biochem.Biophys.Res.Commun., 477, 2016
|
|
4HGX
 
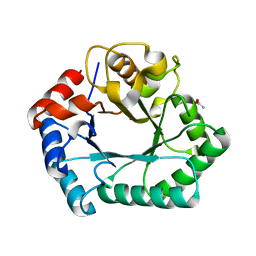 | | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand | | Descriptor: | ACETATE ION, Xylose isomerase domain containing protein, ZINC ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Korzhenevskiy, D.A, Dorovatovsky, P.V, Rakitina, T.V, Lipkin, A.V, Shumilin, I.A, Minor, W, Popov, V.O. | | Deposit date: | 2012-10-09 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand
To be Published
|
|
4HX0
 
 | | Crystal structure of a putative nucleotidyltransferase (TM1012) from Thermotoga maritima at 1.87 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-BUTANEDIOL, Putative nucleotidyltransferase TM1012, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Korzhenevskiy, D.A, Lipkin, A.V, Popov, V.O, Kovalchuk, M.V, Shumilin, I.A, Minor, W, Shabalin, I.G, Golubev, A.M. | | Deposit date: | 2012-11-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of putative nucleotidyltransferase with two MPD molecules in the active site.
To be Published
|
|
5EKC
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Shabalin, I.G, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure of thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+
To Be Published
|
|
5EK6
 
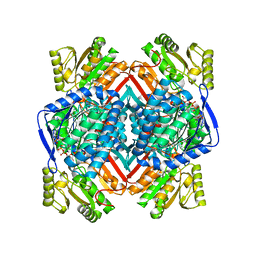 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 complexed with NADP and isobutyraldehyde | | Descriptor: | 2-methylpropanal, Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Polyakov, K.M, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
7ZTQ
 
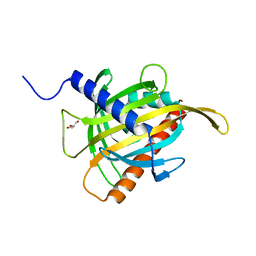 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform | | Descriptor: | Carotenoid-binding protein, GLYCEROL | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
7ZTU
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, D162L mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
7ZVQ
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, S206V mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
