5LJ3
 
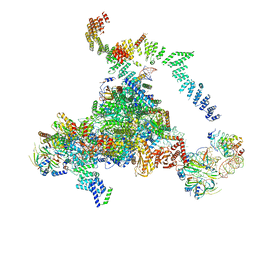 | | Structure of the core of the yeast spliceosome immediately after branching | | 分子名称: | CEF1, CLF1, CWC15, ... | | 著者 | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | 登録日 | 2016-07-17 | | 公開日 | 2016-08-03 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5LSB
 
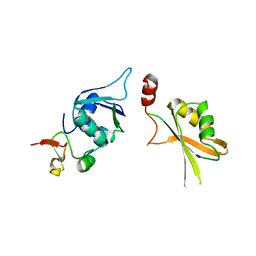 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | 分子名称: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | 著者 | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | 登録日 | 2016-08-24 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
5LSL
 
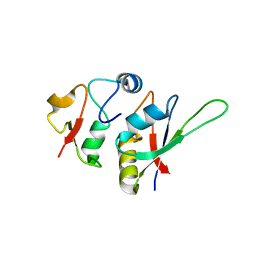 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | 分子名称: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | 著者 | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | 登録日 | 2016-09-02 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | 分子名称: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | 著者 | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | 登録日 | 2016-07-17 | | 公開日 | 2016-08-31 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
1D3B
 
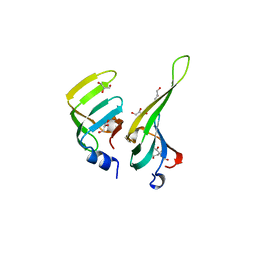 | | CRYSTAL STRUCTURE OF THE D3B SUBCOMPLEX OF THE HUMAN CORE SNRNP DOMAIN AT 2.0A RESOLUTION | | 分子名称: | CITRIC ACID, GLYCEROL, PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN ASSOCIATED PROTEIN B), ... | | 著者 | Kambach, C, Walke, S, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | 登録日 | 1998-12-22 | | 公開日 | 1999-12-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
2BN6
 
 | | P-Element Somatic Inhibitor Protein | | 分子名称: | PSI | | 著者 | Ignjatovic, T, Yang, J.C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | 登録日 | 2005-03-21 | | 公開日 | 2005-07-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
2BN5
 
 | | P-Element Somatic Inhibitor Protein Complex with U1-70k proline-rich peptide | | 分子名称: | PSI, U1 SMALL NUCLEAR RIBONUCLEOPROTEIN 70 KDA | | 著者 | Ignjatovic, T, Yang, J.-C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | 登録日 | 2005-03-21 | | 公開日 | 2005-07-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
6G90
 
 | | Prespliceosome structure provides insight into spliceosome assembly and regulation (map A2) | | 分子名称: | 56 kDa U1 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing factor 39, ... | | 著者 | Plaschka, C, Lin, P.-C, Charenton, C, Nagai, K. | | 登録日 | 2018-04-10 | | 公開日 | 2018-08-22 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Prespliceosome structure provides insights into spliceosome assembly and regulation.
Nature, 559, 2018
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | 分子名称: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | 著者 | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | 登録日 | 2011-04-04 | | 公開日 | 2011-12-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
1A9N
 
 | |
1B34
 
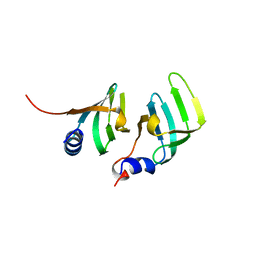 | | CRYSTAL STRUCTURE OF THE D1D2 SUB-COMPLEX FROM THE HUMAN SNRNP CORE DOMAIN | | 分子名称: | PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN SM D1), PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN SM D2) | | 著者 | Walke, S, Young, R.J, Kambach, C, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | 登録日 | 1998-12-17 | | 公開日 | 2000-01-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
1AUD
 
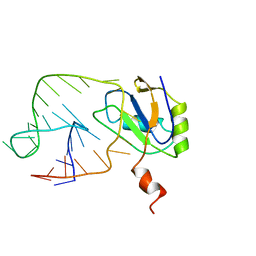 | | U1A-UTRRNA, NMR, 31 STRUCTURES | | 分子名称: | RNA 3UTR, U1A 102 | | 著者 | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | 登録日 | 1997-08-22 | | 公開日 | 1998-02-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of the RNA-binding specificity of human U1A protein.
EMBO J., 16, 1997
|
|
1L9A
 
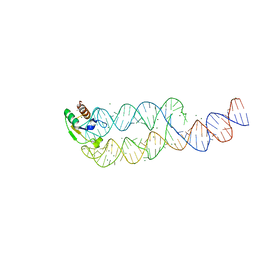 | | CRYSTAL STRUCTURE OF SRP19 IN COMPLEX WITH THE S DOMAIN OF SIGNAL RECOGNITION PARTICLE RNA | | 分子名称: | MAGNESIUM ION, METHYL MERCURY ION, SIGNAL RECOGNITION PARTICLE 19 KDA PROTEIN, ... | | 著者 | Oubridge, C, Kuglstatter, A, Jovine, L, Nagai, K. | | 登録日 | 2002-03-22 | | 公開日 | 2002-06-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle.
Mol.Cell, 9, 2002
|
|
1MFQ
 
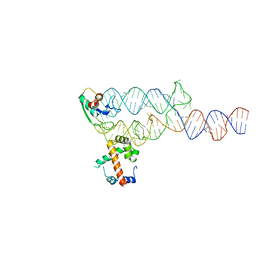 | | Crystal Structure Analysis of a Ternary S-Domain Complex of Human Signal Recognition Particle | | 分子名称: | 7S RNA of human SRP, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Kuglstatter, A, Oubridge, C, Nagai, K. | | 登録日 | 2002-08-13 | | 公開日 | 2002-09-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Induced structural changes of 7SL RNA during the assembly of human
signal recognition particle
Nat.Struct.Biol., 9, 2002
|
|
3ZEF
 
 | | Crystal structure of Prp8:Aar2 complex: second crystal form at 3.1 Angstrom resolution | | 分子名称: | A1 CISTRON-SPLICING FACTOR AAR2, PRE-MRNA-SPLICING FACTOR 8 | | 著者 | Galej, W.P, Oubridge, C, Newman, A.J, Nagai, K. | | 登録日 | 2012-12-05 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of Prp8 Reveals Active Site Cavity of the Spliceosome
Nature, 493, 2013
|
|
4BGD
 
 | | Crystal structure of Brr2 in complex with the Jab1/MPN domain of Prp8 | | 分子名称: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Nguyen, T.H.D, Li, J, Nagai, K. | | 登録日 | 2013-03-25 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis of Brr2-Prp8 Interactions and Implications for U5 Snrnp Biogenesis and the Spliceosome Active Site
Structure, 21, 2013
|
|
5XYO
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G mutant | | 分子名称: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYP
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122R mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYT
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., H130Y mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SULFATE ION | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72. | | 分子名称: | CHLORIDE ION, Endotype 6-aminohexanoat-oligomer hydrolase, GLYCEROL, ... | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-07 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYS
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122V mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y0L
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G/H130Y mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SODIUM ION, ... | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-18 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.385 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y0M
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D36A/D122G/H130Y/E263Q mutant | | 分子名称: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-18 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYQ
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122K mutant | | 分子名称: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | 著者 | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | 登録日 | 2017-07-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
7YU1
 
 | |
