6MEZ
 
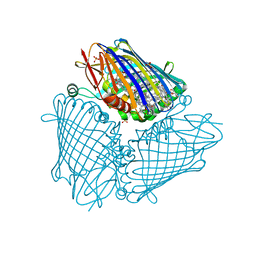 | | X-ray structure of the Fenna-Matthews-Olsen antenna complex from Prosthecochloris aestuarii | | 分子名称: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein, SULFATE ION | | 著者 | Selvaraj, B, Lu, X, Cuneo, M.J, Myles, D.A.A. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Neutron and X-ray analysis of the Fenna-Matthews-Olson photosynthetic antenna complex from Prosthecochloris aestuarii.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
1GKT
 
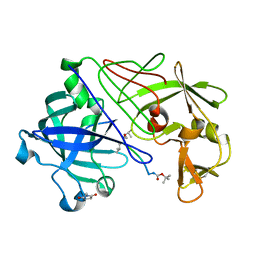 | | Neutron Laue diffraction structure of endothiapepsin complexed with transition state analogue inhibitor H261 | | 分子名称: | ENDOTHIAPEPSIN, INHIBITOR, H261 | | 著者 | Coates, L, Erskine, P.T, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | 登録日 | 2001-08-20 | | 公開日 | 2001-11-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | NEUTRON DIFFRACTION (2.1 Å) | | 主引用文献 | A Neutron Laue Diffraction Study of Endothiapepsin: Implications for the Aspartic Proteinase Mechanism
Biochemistry, 40, 2001
|
|
1XQN
 
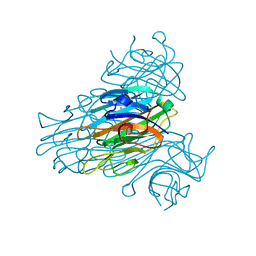 | | The 15k neutron structure of saccharide-free concanavalin A | | 分子名称: | CALCIUM ION, Concanavalin A, MANGANESE (II) ION | | 著者 | Blakeley, M.P, Kalb-Gilboa, A.J, Helliwell, J.R, Myles, D.A.A. | | 登録日 | 2004-10-13 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (2.5 Å) | | 主引用文献 | The 15-K neutron structure of saccharide-free concanavalin A
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1OEW
 
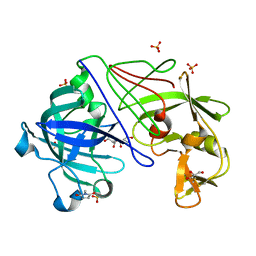 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | 分子名称: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | 著者 | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | 登録日 | 2003-03-31 | | 公開日 | 2003-04-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OEX
 
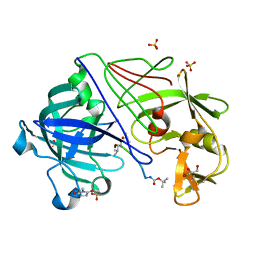 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | 分子名称: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | 著者 | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | 登録日 | 2003-03-31 | | 公開日 | 2003-04-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | 分子名称: | CHLORIDE ION, Endolysin | | 著者 | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | 登録日 | 2017-05-01 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | NEUTRON DIFFRACTION (2.2 Å) | | 主引用文献 | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VNR
 
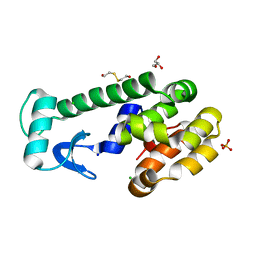 | | X-ray structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | 著者 | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | 登録日 | 2017-05-01 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.631 Å) | | 主引用文献 | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5EAJ
 
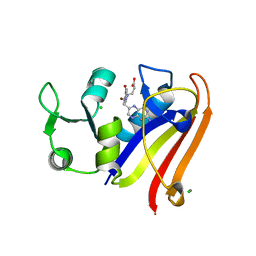 | |
1QTJ
 
 | |
1YRC
 
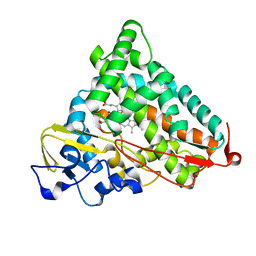 | | X-ray Crystal Structure of hydrogenated Cytochrome P450cam | | 分子名称: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | 著者 | Meilleur, F, Dauvergne, M.-T, Schlichting, I, Myles, D.A.A. | | 登録日 | 2005-02-03 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Production and X-ray crystallographic analysis of fully deuterated cytochrome P450cam.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5HM4
 
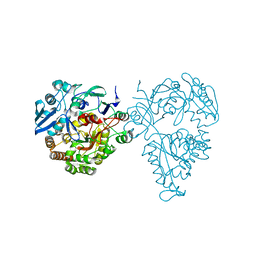 | | Crystal structure of oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.0 A resolution | | 分子名称: | CALCIUM ION, Mannoside ABC transport system, sugar-binding protein | | 著者 | Lu, X, Ghimire-Rijal, S, Myles, D.A.A, Cuneo, M.J. | | 登録日 | 2016-01-15 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Periplasmic Binding Protein Dimer Has a Second Allosteric Event Tied to Ligand Binding.
Biochemistry, 56, 2017
|
|
5H8Z
 
 | |
2AX2
 
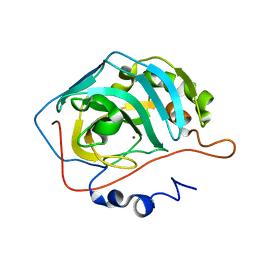 | | Production and X-ray crystallographic analysis of fully deuterated human carbonic anhydrase II | | 分子名称: | Carbonic anhydrase II, ZINC ION | | 著者 | Budayova-Spano, M, Fisher, S.Z, Dauvergne, M.T, Silverman, D.N, Myles, D.A.A, McKenna, R.M. | | 登録日 | 2005-09-02 | | 公開日 | 2006-01-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Production and X-ray crystallographic analysis of fully deuterated human carbonic anhydrase II.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1YRD
 
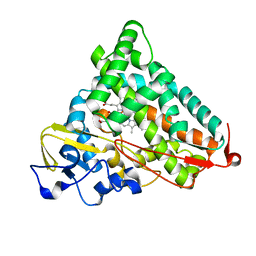 | | X-ray crystal structure of PERDEUTERATED Cytochrome P450cam | | 分子名称: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | 著者 | Meilleur, F, Dauvergne, M.-T, Schlichting, I, Myles, D.A.A. | | 登録日 | 2005-02-03 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Production and X-ray crystallographic analysis of fully deuterated cytochrome P450cam.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6NAF
 
 | | De novo designed homo-trimeric amantadine-binding protein | | 分子名称: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | 著者 | Selvaraj, B, Park, J, Cuneo, M.J, Myles, D.A.A, Baker, D. | | 登録日 | 2018-12-05 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | NEUTRON DIFFRACTION (1.923 Å), X-RAY DIFFRACTION | | 主引用文献 | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
3RZ6
 
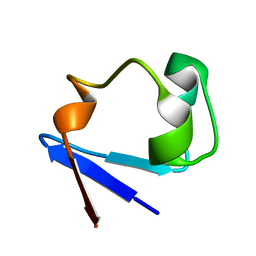 | | Neutron structure of perdeuterated rubredoxin using 40 hours 1st pass data | | 分子名称: | FE (III) ION, Rubredoxin | | 著者 | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | 登録日 | 2011-05-11 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | NEUTRON DIFFRACTION (1.75 Å) | | 主引用文献 | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RZT
 
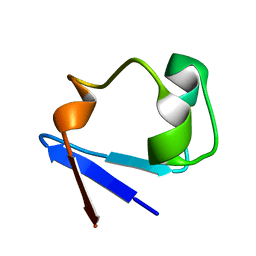 | | Neutron structure of perdeuterated rubredoxin using rapid (14 hours) data | | 分子名称: | FE (III) ION, Rubredoxin | | 著者 | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | 登録日 | 2011-05-12 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | NEUTRON DIFFRACTION (1.7504 Å) | | 主引用文献 | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RYG
 
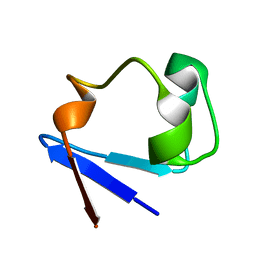 | | 128 hours neutron structure of perdeuterated rubredoxin | | 分子名称: | FE (III) ION, Rubredoxin | | 著者 | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | 登録日 | 2011-05-11 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | NEUTRON DIFFRACTION (1.75 Å) | | 主引用文献 | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SS2
 
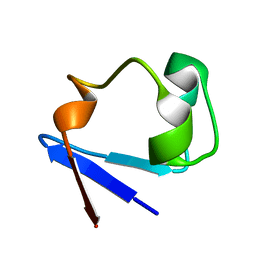 | | Neutron structure of perdeuterated rubredoxin using 48 hours 3rd pass data | | 分子名称: | FE (III) ION, Rubredoxin | | 著者 | Munshi, P, Chung, C.-L, Blakeley, M.P, Weiss, K.L, Myles, D.A.A, Meilleur, F. | | 登録日 | 2011-07-07 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | NEUTRON DIFFRACTION (1.75 Å) | | 主引用文献 | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5UJX
 
 | |
7JOR
 
 | | Neutron structure of ferric Dehaloperoxidase B | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Carey, L.M, Ghiladi, R.A, Meilleur, F, Myles, D.A.A. | | 登録日 | 2020-08-07 | | 公開日 | 2021-09-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (2.05 Å) | | 主引用文献 | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
6DTQ
 
 | | Maltose bound T. maritima MalE3 | | 分子名称: | MAGNESIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE3 | | 著者 | Cuneo, M.J, Shukla, S. | | 登録日 | 2018-06-18 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTS
 
 | | Maltotetraose bound T. maritima MalE2 | | 分子名称: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | 著者 | Cuneo, M.J, Shukla, S. | | 登録日 | 2018-06-18 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTR
 
 | | Apo T. maritima MalE3 | | 分子名称: | SULFATE ION, maltose-binding protein MalE3 | | 著者 | Cuneo, M.J, Shukla, S. | | 登録日 | 2018-06-18 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTT
 
 | | Apo T. maritima MalE2 | | 分子名称: | maltose-binding protein MalE2 | | 著者 | Cuneo, M.J, Shukla, S. | | 登録日 | 2018-06-18 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
