2RVQ
 
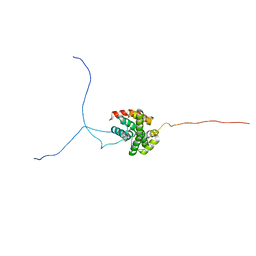 | | Solution structure of the isolated histone H2A-H2B heterodimer | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J | | Authors: | Moriwaki, Y, Yamane, T, Ohtomo, H, Ikeguchi, M, Kurita, J, Sato, M, Nagadoi, A, Shimojo, H, Nishimura, Y. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the isolated histone H2A-H2B heterodimer
Sci Rep, 6, 2016
|
|
3QUG
 
 | | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Gallium-porphyrin | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING GA, ... | | Authors: | Moriwaki, Y, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-02-24 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of recognition of antibacterial porphyrins by heme-transporter IsdH-NEAT3 of Staphylococcus aureus.
Biochemistry, 50, 2011
|
|
3QUH
 
 | |
2AMG
 
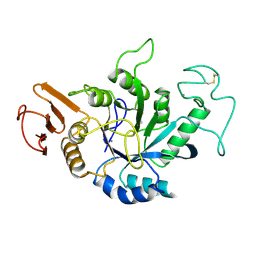 | | STRUCTURE OF HYDROLASE (GLYCOSIDASE) | | Descriptor: | 1,4-ALPHA-D-GLUCAN MALTOTETRAHYDROLASE, CALCIUM ION | | Authors: | Morishita, Y, Hasegawa, K, Matsuura, Y, Kubota, M, Sakai, S, Katsube, Y. | | Deposit date: | 1996-12-23 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a maltotetraose-forming exo-amylase from Pseudomonas stutzeri.
J.Mol.Biol., 267, 1997
|
|
5B5J
 
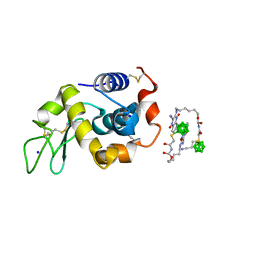 | | Hen egg white lysozyme with boron tracedrug UTX-97 | | Descriptor: | 2-cyano-3-((6-(((2-((2-cyanoethyl)(borocaptate-10B)sulfonio)acetyl)carbamoyl)oxy)hexyl)amino)quinoxaline 1,4-dioxide, Lysozyme C, SODIUM ION | | Authors: | Morimoto, Y. | | Deposit date: | 2016-05-11 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural Insight Into Protein Binding of Boron Tracedrug UTX-97 Revealed by the Co-Crystal Structure With Lysozyme at 1.26 angstrom Resolution.
J Pharm Sci, 105, 2016
|
|
2CYM
 
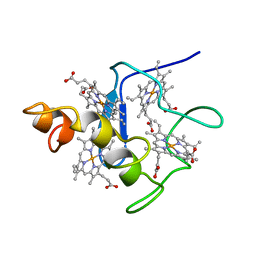 | | EFFECTS OF AMINO ACID SUBSTITUTION ON THREE-DIMENSIONAL STRUCTURE: AN X-RAY ANALYSIS OF CYTOCHROME C3 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH AT 2 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Morimoto, Y, Tani, T, Okumura, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1993-09-29 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of amino acid substitution on three-dimensional structure: an X-ray analysis of cytochrome c3 from Desulfovibrio vulgaris Hildenborough at 2 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
3AB5
 
 | |
5EVY
 
 | | Salicylate hydroxylase substrate complex | | Descriptor: | 2-HYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Salicylate hydroxylase | | Authors: | Morimoto, Y, Uemura, T. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 angstrom resolution
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2E77
 
 | | Crystal structure of L-lactate oxidase with pyruvate complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lactate oxidase, PYRUVIC ACID | | Authors: | Morimoto, Y. | | Deposit date: | 2007-01-06 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study on the interaction of L-lactate oxidase with pyruvate at 1.9 Angstrom resolution.
Biochem.Biophys.Res.Commun., 358, 2007
|
|
2DU2
 
 | | Crystal Structure Analysis of the L-Lactate Oxidase | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Morimoto, Y. | | Deposit date: | 2006-07-19 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of L-lactate oxidase from Aerococcus viridans at 2.1A resolution reveals the mechanism of strict substrate recognition
Biochem.Biophys.Res.Commun., 350, 2006
|
|
6IHX
 
 | | Crystal Structure Analysis of bovine Hemoglobin modified by SNP | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Kihira, K, Morita, Y, Yamada, T, Kureishi, M, Komatsu, T. | | Deposit date: | 2018-10-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Quaternary Structure Analysis of a Hemoglobin Core in Hemoglobin-Albumin Cluster.
J Phys Chem B, 122, 2018
|
|
6II1
 
 | | Crystal Structure Analysis of CO form hemoglobin from Bos taurus | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Kihira, K, Morita, Y, Yamada, T, Kureishi, M, Komatsu, T. | | Deposit date: | 2018-10-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Quaternary Structure Analysis of a Hemoglobin Core in Hemoglobin-Albumin Cluster.
J Phys Chem B, 122, 2018
|
|
5AZQ
 
 | | Crystal structure of cyano-cobalt(III) tetradehydrocorrin in the heme pocket of horse heart myoglobin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, CYANIDE ION, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Inoue, T, Hayashi, T. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures and Coordination Behavior of Aqua- and Cyano-Co(III) Tetradehydrocorrins in the Heme Pocket of Myoglobin
Inorg.Chem., 55, 2016
|
|
5AZR
 
 | | Crystal structure of aqua-cobalt(III) tetradehydrocorrin in the heme pocket of horse heart myoglobin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, GLYCEROL, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Inoue, T, Hayashi, T. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures and Coordination Behavior of Aqua- and Cyano-Co(III) Tetradehydrocorrins in the Heme Pocket of Myoglobin
Inorg.Chem., 55, 2016
|
|
2DXM
 
 | | Neutron Structure Analysis of Deoxy Human Hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Morimoto, Y. | | Deposit date: | 2006-08-28 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Protonation states of buried histidine residues in human deoxyhemoglobin revealed by neutron crystallography.
J.Am.Chem.Soc., 129, 2007
|
|
6KIQ
 
 | | Complex of yeast cytoplasmic dynein MTBD-High and MT with DTT | | Descriptor: | Alpha tubulin, Dynein heavy chain, cytoplasmic, ... | | Authors: | Komori, Y, Nishida, N, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6KIO
 
 | | Complex of yeast cytoplasmic dynein MTBD-High and MT without DTT | | Descriptor: | Dynein heavy chain, cytoplasmic, Tubulin alpha-1A chain, ... | | Authors: | Komori, Y, Nishida, N, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-03-04 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
7F22
 
 | | L-lactate oxidase with pyruvate | | Descriptor: | FLAVIN MONONUCLEOTIDE, L-lactate oxidase, PYRUVIC ACID | | Authors: | Morimoto, Y, Inaka, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Dynamic interactions in the l-lactate oxidase active site facilitate substrate binding at pH4.5.
Biochem.Biophys.Res.Commun., 568, 2021
|
|
7F20
 
 | | L-lactate oxidase with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, FLAVIN MONONUCLEOTIDE, L-lactate oxidase | | Authors: | Morimoto, Y, Inaka, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dynamic interactions in the l-lactate oxidase active site facilitate substrate binding at pH4.5.
Biochem.Biophys.Res.Commun., 568, 2021
|
|
7F21
 
 | | L-lactate oxidase with D-lactate | | Descriptor: | FLAVIN MONONUCLEOTIDE, L-lactate oxidase, LACTIC ACID | | Authors: | Morimoto, Y, Inaka, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Dynamic interactions in the l-lactate oxidase active site facilitate substrate binding at pH4.5.
Biochem.Biophys.Res.Commun., 568, 2021
|
|
7F1Y
 
 | | L-lactate oxidase without substrate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Morimoto, Y, Inaka, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Dynamic interactions in the l-lactate oxidase active site facilitate substrate binding at pH4.5.
Biochem.Biophys.Res.Commun., 568, 2021
|
|
1CCR
 
 | | STRUCTURE OF RICE FERRICYTOCHROME C AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Ochi, H, Hata, Y, Tanaka, N, Kakudo, M, Sakurai, T, Aihara, S, Morita, Y. | | Deposit date: | 1983-03-14 | | Release date: | 1983-04-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of rice ferricytochrome c at 2.0 A resolution.
J.Mol.Biol., 166, 1983
|
|
8XI6
 
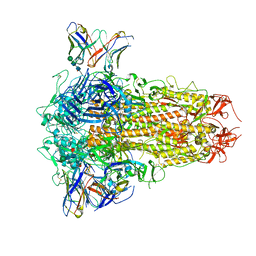 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 2024
|
|
1JJU
 
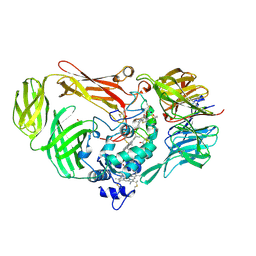 | | Structure of a Quinohemoprotein Amine Dehydrogenase with a Unique Redox Cofactor and Highly Unusual Crosslinking | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, QUINOHEMOPROTEIN AMINE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Datta, S, Mori, Y, Takagi, K, Kawaguchi, K, Chen, Z.-W, Kano, K, Ikeda, T, Okajima, T, Kuroda, S, Tanizawa, K, Mathews, F.S. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8H3M
 
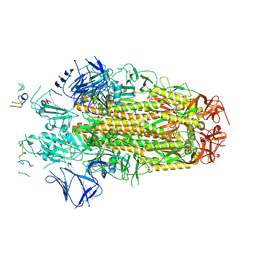 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
