2DKD
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the product complex | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-galactopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2ED4
 
 | | Crystal structure of flavin reductase HpaC complexed with FAD and NAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygenae | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2E6E
 
 | |
2E6B
 
 | |
2E69
 
 | |
2E6G
 
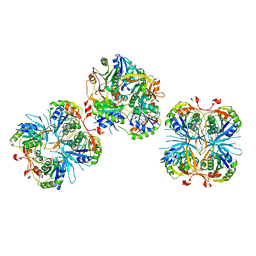 | |
2E6C
 
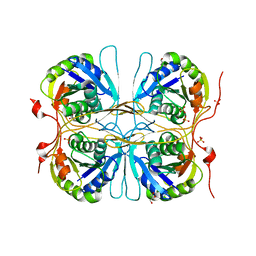 | |
2ECU
 
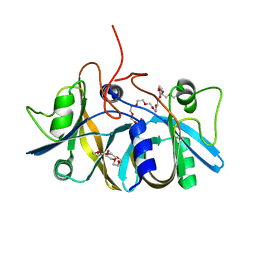 | | Crystal structure of flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DODECAETHYLENE GLYCOL, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygnease | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2E6H
 
 | |
2DKA
 
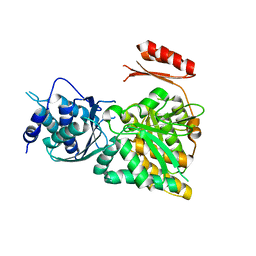 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the apo-form | | Descriptor: | Phosphoacetylglucosamine mutase | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
2CVO
 
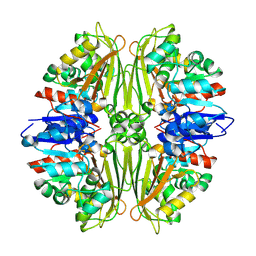 | | Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa) | | Descriptor: | putative Semialdehyde dehydrogenase | | Authors: | Nonaka, T, Kita, A, Miura-Ohnuma, J, Katoh, E, Inagaki, N, Yamazaki, T, Miki, K. | | Deposit date: | 2005-06-10 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa)
Proteins, 61, 2005
|
|
2D2F
 
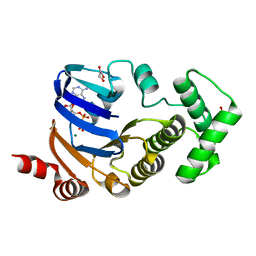 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
3WJW
 
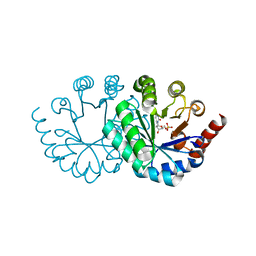 | | Wild-type orotidine 5'-monophosphate decarboxylase from M. thermoautotrophicus complexed with 6-methyl-UMP | | Descriptor: | 6-methyluridine 5'-(dihydrogen phosphate), Orotidine 5'-phosphate decarboxylase | | Authors: | Fujihashi, M, Kuroda, S, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
3WJX
 
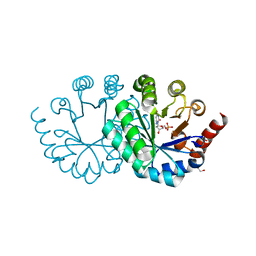 | | Wild-type orotidine 5'-monophosphate decarboxylase from M. thermoautotrophicus complexed with 6-amino-UMP | | Descriptor: | 6-AMINOURIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fujihashi, M, Kuroda, S, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
3WMQ
 
 | | Crystal structure of the complex between SLL-2 and GalNAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kita, A, Jimbo, M, Sakai, R, Morimoto, Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a symbiosis-related lectin from octocoral.
Glycobiology, 25, 2015
|
|
3WJY
 
 | |
3WDK
 
 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase complexed with reaction intermediate | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, 5'-O-[(S)-hydroxy{[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]oxy}phosphoryl]adenosine, CITRATE ANION | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
3WJP
 
 | | Crystal structure of the HypE CA form | | Descriptor: | BENZAMIDINE, GLYCEROL, Hydrogenase expression/formation protein HypE, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WK1
 
 | |
3WJR
 
 | | crystal structure of HypE in complex with a nucleotide | | Descriptor: | ADENOSINE MONOPHOSPHATE, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WJZ
 
 | | Orotidine 5'-monophosphate decarboxylase D75N mutant from M. thermoautotrophicus complexed with 6-amino-UMP | | Descriptor: | 6-AMINOURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujihashi, M, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
3WDM
 
 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase from Thermococcus kodakarensis | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, ADENOSINE | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
3WK2
 
 | |
3WMP
 
 | | Crystal structure of SLL-2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kita, A, Jimbo, M, Sakai, R, Morimoto, Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a symbiosis-related lectin from octocoral.
Glycobiology, 25, 2015
|
|
