7VLC
 
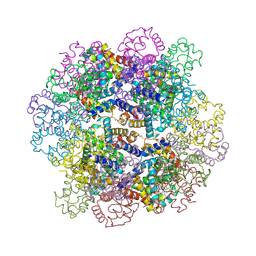 | | Oxy-deoxy intermediate of V2 hemoglobin at 78% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLD
 
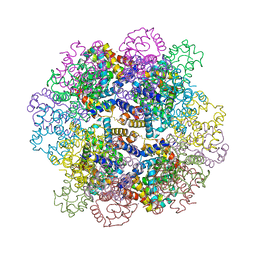 | | Oxy-deoxy intermediate of V2 hemoglobin at 69% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLF
 
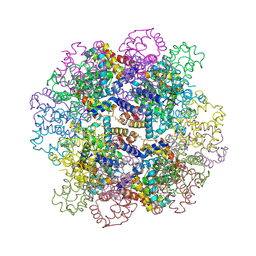 | | Oxy-deoxy intermediate of V2 hemoglobin at 26% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLE
 
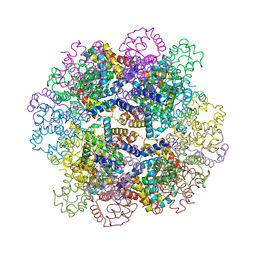 | | Oxy-deoxy intermediate of V2 hemoglobin at 55% oxygen saturation | | Descriptor: | Extracellular A1 globin, Extracellular A2 globin, Extracellular B1 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
5WQR
 
 | | High resolution structure of high-potential iron-sulfur protein in the reduced state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
5WQQ
 
 | | High resolution structure of high-potential iron-sulfur protein in the oxidized state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
5HEE
 
 | | Crystal structure of the TK2203 protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, TK2203 protein, ... | | Authors: | Nishitani, Y, Miki, K. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of the TK2203 protein from Thermococcus kodakarensis, a putative extradiol dioxygenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5HWS
 
 | |
5IJA
 
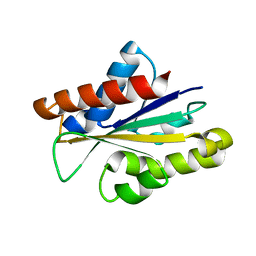 | |
1EYT
 
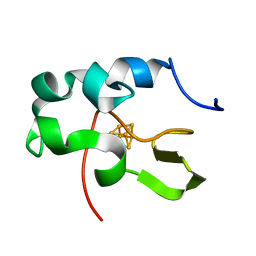 | | CRYSTAL STRUCTURE OF HIGH-POTENTIAL IRON-SULFUR PROTEIN FROM THERMOCHROMATIUM TEPIDUM | | Descriptor: | HIGH-POTENTIAL IRON-SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Nogi, T, Fathir, I, Kobayashi, M, Nozawa, T, Miki, K. | | Deposit date: | 2000-05-08 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of photosynthetic reaction center and high-potential iron-sulfur protein from Thermochromatium tepidum: thermostability and electron transfer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F75
 
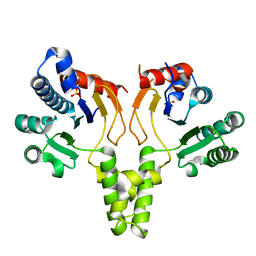 | | CRYSTAL STRUCTURE OF UNDECAPRENYL DIPHOSPHATE SYNTHASE FROM MICROCOCCUS LUTEUS B-P 26 | | Descriptor: | SULFATE ION, UNDECAPRENYL PYROPHOSPHATE SYNTHETASE | | Authors: | Fujihashi, M, Zhang, Y.-W, Higuchi, Y, Li, X.-Y, Koyama, T, Miki, K. | | Deposit date: | 2000-06-26 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1FVP
 
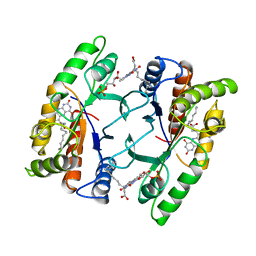 | | FLAVOPROTEIN 390 | | Descriptor: | 6-(3-TETRADECANOIC ACID) FLAVINE MONONUCLEOTIDE, FLAVOPROTEIN 390 | | Authors: | Kita, A, Miki, K. | | Deposit date: | 1995-07-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of flavoprotein FP390 from a luminescent bacterium Photobacterium phosphoreum refined at 2.7 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1MPY
 
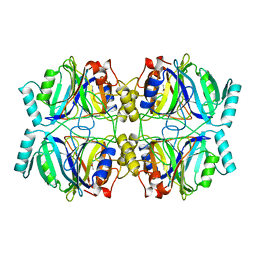 | | STRUCTURE OF CATECHOL 2,3-DIOXYGENASE (METAPYROCATECHASE) FROM PSEUDOMONAS PUTIDA MT-2 | | Descriptor: | ACETONE, CATECHOL 2,3-DIOXYGENASE, FE (II) ION | | Authors: | Kita, A, Kita, S, Fujisawa, I, Inaka, K, Ishida, T, Horiike, K, Nozaki, M, Miki, K. | | Deposit date: | 1998-10-20 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2,3-dioxygenase (metapyrocatechase) from Ppseudomonas putida mt-2.
Structure Fold.Des., 7, 1999
|
|
1IUA
 
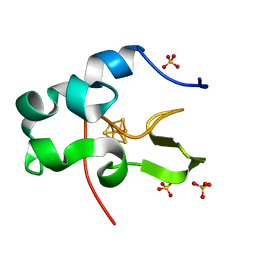 | | Ultra-high resolution structure of HiPIP from Thermochromatium tepidum | | Descriptor: | High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Liu, L, Nogi, T, Kobayashi, M, Nozawa, T, Miki, K. | | Deposit date: | 2002-03-01 | | Release date: | 2002-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Ultrahigh-resolution structure of high-potential iron-sulfur protein from Thermochromatium tepidum.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4GA6
 
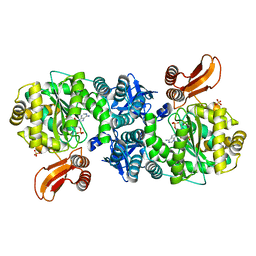 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in complex with substrates | | Descriptor: | ADENOSINE MONOPHOSPHATE, Putative thymidine phosphorylase, SULFATE ION | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
4G9I
 
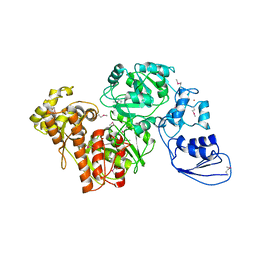 | | Crystal structure of T.kodakarensis HypF | | Descriptor: | Hydrogenase maturation protein HypF, ZINC ION | | Authors: | Tominaga, T, Watanabe, S, Matsumi, R, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-24 | | Release date: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of the [NiFe]-hydrogenase maturation protein HypF from Thermococcus kodakarensis KOD1.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4GA4
 
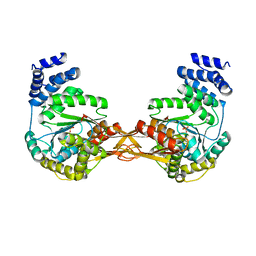 | | Crystal structure of AMP phosphorylase N-terminal deletion mutant | | Descriptor: | PHOSPHATE ION, Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
6JGJ
 
 | | Crystal structure of the F99S/M153T/V163A/E222Q variant of GFP at 0.78 A | | Descriptor: | Green fluorescent protein, MAGNESIUM ION | | Authors: | Takaba, K, Tai, Y, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-02-14 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
6JGH
 
 | | Crystal structure of the F99S/M153T/V163A/T203I variant of GFP at 0.94 A | | Descriptor: | CHLORIDE ION, Green fluorescent protein | | Authors: | Eki, H, Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-02-14 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
4GA5
 
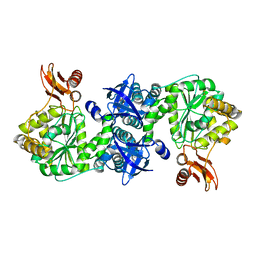 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in the apo-form | | Descriptor: | Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
6JGI
 
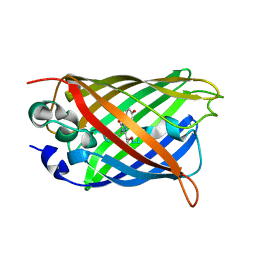 | | Crystal structure of the S65T/F99S/M153T/V163A variant of GFP at 0.85 A | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-02-14 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
5ZUI
 
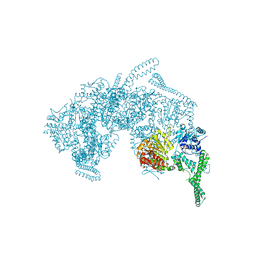 | | Crystal Structure of HSP104 from Chaetomium thermophilum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat Shock Protein 104, SULFATE ION | | Authors: | Hanazono, Y, Inoue, Y, Noguchi, K, Yohda, M, Shinohara, K, Takeda, K, Miki, K. | | Deposit date: | 2018-05-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 2021
|
|
7E6P
 
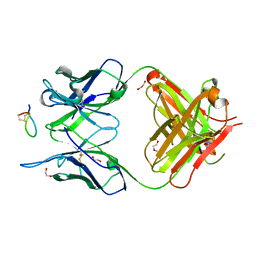 | | Fab-amyloid beta fragment complex | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta fragment with an intramolecular disulfide bond at positions 17 and 28, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kita, A, Irie, K, Irie, Y, Miki, K. | | Deposit date: | 2021-02-23 | | Release date: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of a Conformation-Restricted Amyloid beta Peptide and Immunoreactivity of Its Antibody in Human AD brain.
Acs Chem Neurosci, 12, 2021
|
|
6KL1
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of non-deuterated GFP at pD 8.5 | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Dao, H.A, Miki, K, Takeda, K. | | Deposit date: | 2019-07-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.851 Å) | | Cite: | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
6KL0
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of perdeuterated GFP at pD 7.0 | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-07-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.798 Å) | | Cite: | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
