7ZHS
 
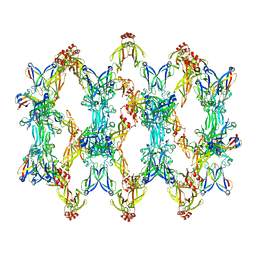 | | 3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F by imposing D5 symmetry | | 分子名称: | Ubiquitin-like protein SMT3,DnaJ homolog subfamily A member 2, ZINC ION | | 著者 | Cuellar, J, Velasco-Carneros, L, Santiago, C, Martin-Benito, J, Valpuesta, J, Muga, A. | | 登録日 | 2022-04-07 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | The self-association equilibrium of DNAJA2 regulates its interaction with unfolded substrate proteins and with Hsc70.
Nat Commun, 14, 2023
|
|
2WFS
 
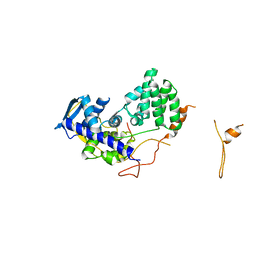 | | Fitting of influenza virus NP structure into the 9-fold symmetryzed cryoEM reconstruction of an active RNP particle. | | 分子名称: | NUCLEOPROTEIN | | 著者 | Coloma, R, Valpuesta, J.M, Arranz, R, Carrascosa, J.L, Ortin, J, Martin-Benito, J. | | 登録日 | 2009-04-15 | | 公開日 | 2009-07-07 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | The Structure of a Biologically Active Influenza Virus Ribonucleoprotein Complex.
Plos Pathog., 5, 2009
|
|
2XQL
 
 | | Fitting of the H2A-H2B histones in the electron microscopy map of the complex Nucleoplasmin:H2A-H2B histones (1:5). | | 分子名称: | HISTONE H2A-IV, HISTONE H2B 5 | | 著者 | Ramos, I, Martin-Benito, J, Finn, R, Bretana, L, Aloria, K, Arizmendi, J.M, Ausio, J, Muga, A, Valpuesta, J.M, Prado, A. | | 登録日 | 2010-09-02 | | 公開日 | 2010-11-03 | | 最終更新日 | 2017-08-30 | | 実験手法 | ELECTRON MICROSCOPY (19.5 Å) | | 主引用文献 | Nucleoplasmin Binds Histone H2A-H2B Dimers Through its Distal Face.
J.Biol.Chem., 285, 2010
|
|
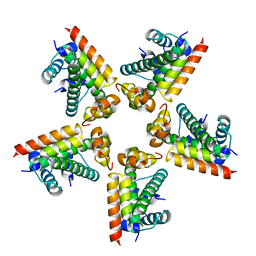
1GWY
 
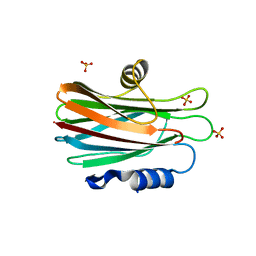 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II | | 分子名称: | STICHOLYSIN II, SULFATE ION | | 著者 | Mancheno, J.M, Martin-Benito, J, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | 登録日 | 2002-03-26 | | 公開日 | 2003-06-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
2XVR
 
 | | Phage T7 empty mature head shell | | 分子名称: | MAJOR CAPSID PROTEIN 10A | | 著者 | Ionel, A, Velazquez-Muriel, J.A, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | 登録日 | 2010-10-28 | | 公開日 | 2010-12-01 | | 最終更新日 | 2018-10-03 | | 実験手法 | ELECTRON MICROSCOPY (10.8 Å) | | 主引用文献 | Molecular Rearrangements Involved in the Capsid Shell Maturation of Bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
4BBL
 
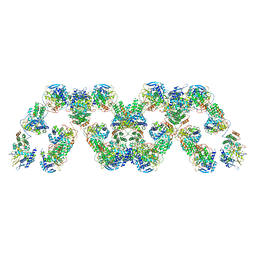 | | Cryo-electron microscopy reconstruction of the helical part of influenza A virus ribonucleoprotein isolated from virions. | | 分子名称: | NUCLEOPROTEIN, RNA | | 著者 | Arranz, R, Coloma, R, Chichon, F.J, Conesa, J.J, Carrascosa, J.L, Valpuesta, J.M, Ortin, J, Martin-Benito, J. | | 登録日 | 2012-09-26 | | 公開日 | 2012-12-05 | | 最終更新日 | 2017-08-23 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | The Structure of Native Influenza Virion Ribonucleoproteins
Science, 338, 2012
|
|
3IZG
 
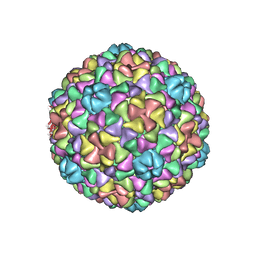 | | Bacteriophage T7 prohead shell EM-derived atomic model | | 分子名称: | Major capsid protein 10A | | 著者 | Ionel, A, Velazquez-Muriel, J.A, Agirrezabala, X, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | 登録日 | 2010-10-27 | | 公開日 | 2010-11-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (10.9 Å) | | 主引用文献 | Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
3J4A
 
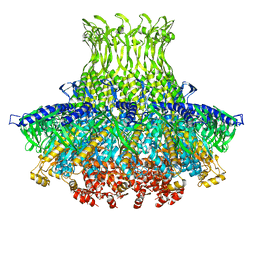 | | Structure of gp8 connector protein | | 分子名称: | Head-to-tail joining protein | | 著者 | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | 登録日 | 2013-07-09 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
3J4B
 
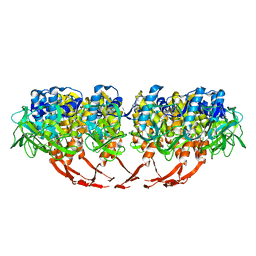 | | Structure of T7 gatekeeper protein (gp11) | | 分子名称: | Tail tubular protein A | | 著者 | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | 登録日 | 2013-07-09 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
4BIJ
 
 | | Threading model of T7 large terminase | | 分子名称: | DNA MATURASE B | | 著者 | Dauden, M.I, Martin-Benito, J, Sanchez-Ferrero, J.C, Pulido-Cid, M, Valpuesta, J.M, Carrascosa, J.L. | | 登録日 | 2013-04-10 | | 公開日 | 2013-05-08 | | 最終更新日 | 2017-08-23 | | 実験手法 | ELECTRON MICROSCOPY (16 Å) | | 主引用文献 | Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T7
J.Biol.Chem., 288, 2013
|
|
4BIL
 
 | | Threading model of the T7 large terminase within the gp8gp19 complex | | 分子名称: | DNA MATURASE B | | 著者 | Dauden, M.I, Martin-Benito, J, Sanchez-Ferrero, J.C, Pulido-Cid, M, Valpuesta, J.M, Carrascosa, J.L. | | 登録日 | 2013-04-10 | | 公開日 | 2013-05-08 | | 最終更新日 | 2017-08-23 | | 実験手法 | ELECTRON MICROSCOPY (29 Å) | | 主引用文献 | Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T7
J.Biol.Chem., 288, 2013
|
|
6I54
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 2. | | 分子名称: | Influenza virus nucleoprotein, Nucleoprotein | | 著者 | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | 登録日 | 2018-11-12 | | 公開日 | 2019-11-13 | | 最終更新日 | 2020-05-27 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6I7M
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 4. | | 分子名称: | Nucleoprotein | | 著者 | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | 登録日 | 2018-11-16 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-08-26 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6I7B
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3. | | 分子名称: | Nucleoprotein | | 著者 | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | 登録日 | 2018-11-16 | | 公開日 | 2020-02-19 | | 最終更新日 | 2020-05-06 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6H9G
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 1. | | 分子名称: | Nucleoprotein, Polypeptide loop | | 著者 | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Munier, S, Carlero, D, Naffakh, N, Ortin, J, Martin-Benito, J. | | 登録日 | 2018-08-03 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-05-06 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6I85
 
 | | Influenza A nucleoprotein docked into the 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 5. | | 分子名称: | Influenza A nucleoprotein, Nucleoprotein | | 著者 | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | 登録日 | 2018-11-19 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-05-06 | | 実験手法 | ELECTRON MICROSCOPY (24 Å) | | 主引用文献 | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
4ICV
 
 | | Ubiquitin-like domain of human tubulin folding cofactor E - crystal form B | | 分子名称: | PRASEODYMIUM ION, Tubulin-specific chaperone E | | 著者 | Janowski, R, Boutin, M, Zabala, J.C, Coll, M. | | 登録日 | 2012-12-11 | | 公開日 | 2014-06-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The structure of the complex between alpha-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism.
J.Cell.Sci., 128, 2015
|
|
4ICU
 
 | | Ubiquitin-like domain of human tubulin folding cofactor E - crystal from A | | 分子名称: | Tubulin-specific chaperone E | | 著者 | Janowski, R, Boutin, M, Zabala, J.C, Coll, M. | | 登録日 | 2012-12-11 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structure of the complex between alpha-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism.
J.Cell.Sci., 128, 2015
|
|
1O72
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II complexed with phosphorylcholine | | 分子名称: | PHOSPHOCHOLINE, STICHOLYSIN II | | 著者 | Mancheno, J.M, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | 登録日 | 2002-10-23 | | 公開日 | 2003-11-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
1O71
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II complexed with glycerol | | 分子名称: | GLYCEROL, STICHOLYSIN II | | 著者 | Mancheno, J.M, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | 登録日 | 2002-10-23 | | 公開日 | 2003-11-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
6I0M
 
 | | Structure of human IMP dehydrogenase, isoform 2, bound to GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Inosine-5'-monophosphate dehydrogenase 2, ... | | 著者 | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | 登録日 | 2018-10-26 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.567 Å) | | 主引用文献 | A Nucleotide-Dependent Conformational Switch Controls the Polymerization of Human IMP Dehydrogenases to Modulate their Catalytic Activity.
J. Mol. Biol., 431, 2019
|
|
6I0O
 
 | | Structure of human IMP dehydrogenase, isoform 2, bound to GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase 2, SULFATE ION | | 著者 | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | 登録日 | 2018-10-26 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.623 Å) | | 主引用文献 | A Nucleotide-Dependent Conformational Switch Controls the Polymerization of Human IMP Dehydrogenases to Modulate their Catalytic Activity.
J. Mol. Biol., 431, 2019
|
|
