3REX
 
 | | Crystal structure of the archaeal asparagine synthetase A complexed with Adenosine monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, AsnS-like asparaginyl-tRNA synthetase related protein, MAGNESIUM ION | | Authors: | Blaise, M, Frechin, M, Charron, C, Roy, H, Sauter, C, Lorber, B, Olieric, V, Kern, D. | | Deposit date: | 2011-04-05 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Archaeal Asparagine Synthetase: Interrelation with Aspartyl-tRNA and Asparaginyl-tRNA Synthetases.
J.Mol.Biol., 412, 2011
|
|
3RL6
 
 | | Crystal structure of the archaeal asparagine synthetase A complexed with L-Asparagine and Adenosine monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE, Archaeal asparagine synthetase A, ... | | Authors: | Blaise, M, Frechin, M, Charron, C, Roy, H, Sauter, C, Lorber, B, Olieric, V, Kern, D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Archaeal Asparagine Synthetase: Interrelation with Aspartyl-tRNA and Asparaginyl-tRNA Synthetases.
J.Mol.Biol., 412, 2011
|
|
1A8H
 
 | | METHIONYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sugiura, I, Nureki, O, Ugaji, Y, Kuwabara, S, Lober, B, Giege, R, Moras, D, Yokoyama, S, Konno, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules.
Structure, 8, 2000
|
|
6Q3T
 
 | | Structure of Protease1 from Pyrococcus horikoshii at room temperature in ChipX microfluidic device | | Descriptor: | Deglycase PH1704 | | Authors: | de Wijn, R, Engilberge, S, Olieric, V, Girard, E, Sauter, C. | | Deposit date: | 2018-12-04 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
4TR7
 
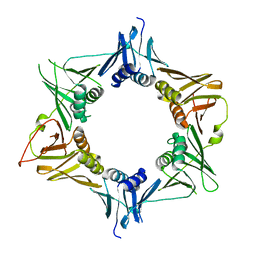 | |
4TR8
 
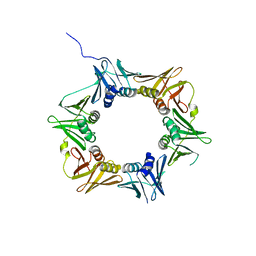 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-15 | | Release date: | 2014-09-10 | | Last modified: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TR6
 
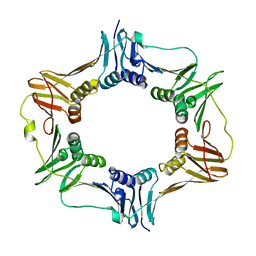 | | Crystal structure of DNA polymerase sliding clamp from Bacillus subtilis | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Burnouf, D, Olieric, V, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-14 | | Release date: | 2014-09-10 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TSZ
 
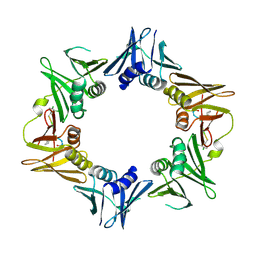 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | Descriptor: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-10 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
2YHT
 
 | |
6FVO
 
 | | Mutant DNA polymerase sliding clamp from Mycobacterium tuberculosis with bound P7 peptide | | Descriptor: | Beta sliding clamp, CALCIUM ION, P7 peptide | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
6FVL
 
 | | DNA polymerase sliding clamp from Escherichia coli with bound P7 peptide | | Descriptor: | Beta sliding clamp, GLYCEROL, P7 peptide, ... | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
6FVM
 
 | | Mutant DNA polymerase sliding clamp from Escherichia coli with bound P7 peptide | | Descriptor: | Beta sliding clamp, CALCIUM ION, GLYCEROL, ... | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
6FVN
 
 | | DNA polymerase sliding clamp from Mycobacterium tuberculosis with bound P7 peptide | | Descriptor: | Beta sliding clamp, P7 peptide | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.142 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
194L
 
 | | THE 1.40 A STRUCTURE OF SPACEHAB-01 HEN EGG WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Vaney, M.C, Maignan, S, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
193L
 
 | | THE 1.33 A STRUCTURE OF TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Vaney, M.C, Maignan, S, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4BS3
 
 | | Bovin insulin structure determined by in situ crystal analysis and sulfur-SAD phasing at room temperature | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Pinker, F, Stirnimann, C, Sauter, C, Olieric, V. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-17 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Chipx: A Novel Microfluidic Chip for Counter- Diffusion Crystallization of Biomolecules and in Situ Crystal Analysis at Room Temperature
J.Cryst.Growth, 13, 2013
|
|
3P8Y
 
 | |
3P8T
 
 | | Crystal structure of the archaeal asparagine synthetase A | | Descriptor: | AsnS-like asparaginyl-tRNA synthetase related protein | | Authors: | Blaise, M, Kern, D. | | Deposit date: | 2010-10-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Archaeal Asparagine Synthetase: Interrelation with Aspartyl-tRNA and Asparaginyl-tRNA Synthetases.
J.Mol.Biol., 412, 2011
|
|
3P8V
 
 | |
6GZP
 
 | | Llama nanobody PorM_02 structure determined at room temperature by in-situ diffraction in ChipX microfluidic device | | Descriptor: | Nanobody | | Authors: | Roche, J, Gaubert, A, Desmyter, A, De Wijn, R, Sauter, C, Roussel, A. | | Deposit date: | 2018-07-04 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6HW1
 
 | | ROOM TEMPERATURE STRUCTURE OF LIPASE FROM T. LANUGINOSA AT 2.5 A RESOLUTION IN CHIPX MICROFLUIDIC DEVICE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Fernadez-Penas, R, Martinez-Rodriguez, S, Verdugo-Escamilla, C. | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
7OQX
 
 | | Crystal structure of a psychrophilic CCA-adding enzyme in complex with CMPcPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ACETATE ION, CCA-adding enzyme, ... | | Authors: | Rollet, K, de Wijn, R, Bluhm, A, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus .
Comput Struct Biotechnol J, 19, 2021
|
|
7OTL
 
 | | Structure of a psychrophilic CCA-adding enzyme crystallized by counter-diffusion | | Descriptor: | CCA-adding enzyme, SULFATE ION | | Authors: | Rollet, K, de Wijn, R, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2021-06-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus .
Comput Struct Biotechnol J, 19, 2021
|
|
3VU8
 
 | | Metionyl-tRNA synthetase from Thermus thermophilus complexed with methionyl-adenylate analogue | | Descriptor: | Methionine--tRNA ligase, N-[METHIONYL]-N'-[ADENOSYL]-DIAMINOSULFONE, ZINC ION | | Authors: | Konno, M, Kato-Murayama, M, Toma-Fukai, S, Uchikawa, E, Nureki, O, Yokoyama, S. | | Deposit date: | 2012-06-22 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The modeling of structures of specific conformation of homosysteine-AMP leading to thiolactone-formation on class Ia aminoacyl-tRNA synthetases
To be Published
|
|
6QXN
 
 | | Crystal structure of the CCA-adding enzyme of a psychrophilic organism in complex with CTP | | Descriptor: | ACETATE ION, CCA-adding enzyme, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | de Wijn, R, Rollet, K, Bluhm, A, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus
Comput Struct Biotechnol J, 2021
|
|
