8H04
 
 | |
7YNQ
 
 | |
7YNT
 
 | |
7YNM
 
 | | ThT-bound alpha-synuclein fibrils conformation 1 | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Alpha-synuclein | | Authors: | Tao, Y.Q, Zhao, Q.Y, Liu, C, Li, D. | | Deposit date: | 2022-07-31 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | ThT-bound alpha-synuclein fibrils conformation 1
To Be Published
|
|
7YNR
 
 | |
7YNL
 
 | | EB-bound alpha-synuclein fibrils | | Descriptor: | 4-azanyl-6-[[4-[4-[(~{E})-(8-azanyl-1-oxidanyl-5,7-disulfo-naphthalen-2-yl)diazenyl]-3-methyl-phenyl]-2-methyl-phenyl]diazenyl]-5-oxidanyl-naphthalene-1,3-disulfonic acid, Alpha-synuclein | | Authors: | Tao, Y.Q, Zhao, Q.Y, Liu, C, Li, D. | | Deposit date: | 2022-07-31 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | EB-bound alpha-synuclein fibrils
To Be Published
|
|
7YNP
 
 | | BF227-bound alpha-synuclein fibrils | | Descriptor: | 5-[(~{E})-2-(6-methoxy-1,3-benzoxazol-2-yl)ethenyl]-~{N},~{N}-dimethyl-1,3-thiazol-2-amine, Alpha-synuclein | | Authors: | Tao, Y.Q, Zhao, Q.Y, Liu, C, Li, D. | | Deposit date: | 2022-07-31 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | BF227-bound alpha-synuclein fibrils
To Be Published
|
|
7YNN
 
 | | ThT-bound alpha-synuclein fibrils conformation 2 | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Alpha-synuclein | | Authors: | Tao, Y.Q, Zhao, Q.Y, Liu, C, Li, D. | | Deposit date: | 2022-07-31 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | ThT-bound alpha-synuclein fibrils conformation 2
To Be Published
|
|
7YNO
 
 | |
7YNS
 
 | |
7YNG
 
 | |
7YNF
 
 | | CCA-bound alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, copper;trisodium;18-(2-carboxylatoethyl)-20-(carboxylatomethyl)-12-ethenyl-7-ethyl-3,8,13,17-tetramethyl-17,18-dihydroporphyrin-21,23-diide-2-carboxylate | | Authors: | Tao, Y.Q, Zhao, Q.Y, Liu, C, Li, D. | | Deposit date: | 2022-07-30 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | CCA-bound alpha-synuclein fibrils
To Be Published
|
|
7C1D
 
 | |
5W74
 
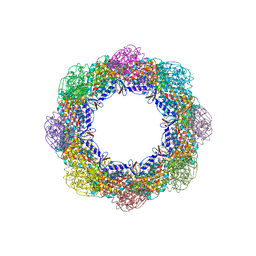 | | Crystal Structure of the Group II Chaperonin from Methanococcus Maripaludis D386ADeltaLid Mutant in the Open, ADP-Bound State | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION | | Authors: | Dalton, K.M, Lopez, T, Liu, C, Ralston, C.Y, Pereira, J.H, Chartron, J.W, McAndrew, R.P, Douglas, N.R, Adams, P.D, Pande, V.S, Frydman, J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The Conformational Cycle of the Group II Chaperonin Termini
To Be Published
|
|
5W79
 
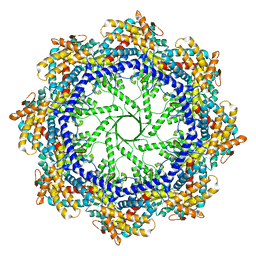 | | Crystal Structure of the Group II Chaperonin from Methanococcus Maripaludis, Cysteine-less mutant in the Apo State | | Descriptor: | Chaperonin, SULFATE ION | | Authors: | Dalton, K.M, Lopez, T, Liu, C, Ralston, C.Y, Pereira, J.H, Chartron, J.W, McAndrew, R.P, Douglas, N.R, Adams, P.D, Pande, V.S, Frydman, J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-20 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (3.122 Å) | | Cite: | The Conformational Cycle of the Group II Chaperonin Termini
To Be Published
|
|
5WSK
 
 | |
8J29
 
 | |
8J2A
 
 | |
8J2D
 
 | |
8J2C
 
 | |
8J2E
 
 | |
8J2B
 
 | |
8J7N
 
 | |
8J7P
 
 | |
8WCL
 
 | | FCP pentamer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Liu, C, Shen, J.-R, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 2024
|
|
