1Y8Q
 
 | | SUMO E1 ACTIVATING ENZYME SAE1-SAE2-MG-ATP COMPLEX | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-like 1 activating enzyme E1A, ... | | 著者 | Lois, L.M, Lima, C.D. | | 登録日 | 2004-12-13 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1
Embo J., 24, 2005
|
|
1Z3C
 
 | | Encephalitozooan cuniculi mRNA Cap (Guanine-N7) Methyltransferasein complexed with AzoAdoMet | | 分子名称: | S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE, mRNA CAPPING ENZYME | | 著者 | Hausmann, S, Zhang, S, Fabrega, C, Schneller, S.W, Lima, C.D, Shuman, S. | | 登録日 | 2005-03-11 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Encephalitozoon cuniculi mRNA cap (guanine N-7) methyltransferase: methyl acceptor specificity, inhibition BY S-adenosylmethionine analogs, and structure-guided mutational analysis.
J.Biol.Chem., 280, 2005
|
|
1Z9D
 
 | | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes | | 分子名称: | SULFATE ION, uridylate kinase | | 著者 | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2005-04-01 | | 公開日 | 2005-04-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes
To be Published
|
|
1Z5S
 
 | | Crystal structure of a complex between UBC9, SUMO-1, RANGAP1 and NUP358/RANBP2 | | 分子名称: | Ran GTPase-activating protein 1, Ran-binding protein 2, Ubiquitin-conjugating enzyme E2 I, ... | | 著者 | Reverter, D, Lima, C.D. | | 登録日 | 2005-03-19 | | 公開日 | 2005-06-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex.
Nature, 435, 2005
|
|
2A8X
 
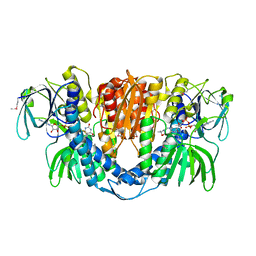 | | Crystal Structure of Lipoamide Dehydrogenase from Mycobacterium tuberculosis | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Rajashankar, K.R, Bryk, R, Kniewel, R, Buglino, J.A, Nathan, C.F, Lima, C.D. | | 登録日 | 2005-07-10 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure and functional analysis of lipoamide dehydrogenase from Mycobacterium tuberculosis
J.Biol.Chem., 280, 2005
|
|
2AP9
 
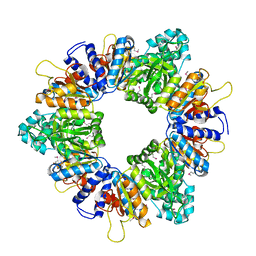 | | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551 | | 分子名称: | MAGNESIUM ION, NICKEL (II) ION, acetylglutamate kinase | | 著者 | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2005-08-15 | | 公開日 | 2005-08-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551
To be Published
|
|
2AR0
 
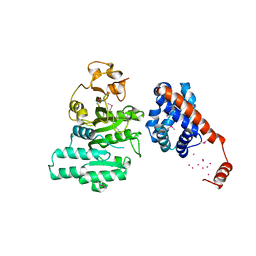 | |
8DAR
 
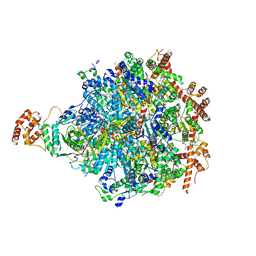 | |
8DAU
 
 | |
8DAW
 
 | |
8DAS
 
 | |
8DAT
 
 | |
8DAV
 
 | |
2HVQ
 
 | | Structure of Adenylated full-length T4 RNA Ligase 2 | | 分子名称: | Hypothetical 37.6 kDa protein in Gp24-hoc intergenic region, MAGNESIUM ION | | 著者 | Nandakumar, J, Lima, C.D. | | 登録日 | 2006-07-30 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVS
 
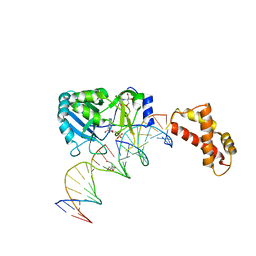 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 2'-deoxyribonucleotide at the nick | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | 著者 | Nandakumar, J, Lima, C.D. | | 登録日 | 2006-07-30 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVR
 
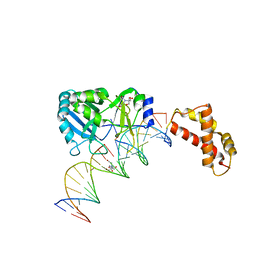 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 3'-deoxyribonucleotide at the nick | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | 著者 | Nandakumar, J, Lima, C.D. | | 登録日 | 2006-07-30 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2IO2
 
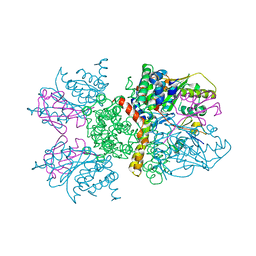 | |
2IO3
 
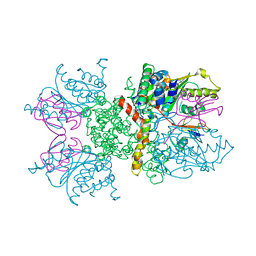 | |
2IO0
 
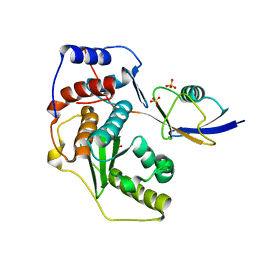 | |
2IO1
 
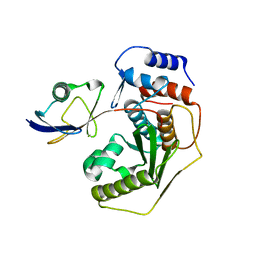 | |
1P16
 
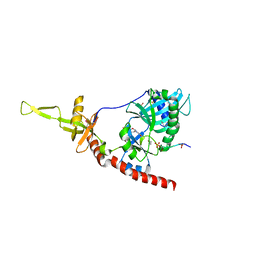 | | Structure of an mRNA capping enzyme bound to the phosphorylated carboxyl-terminal domain of RNA polymerase II | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | 著者 | Fabrega, C, Shen, V, Shuman, S, Lima, C.D. | | 登録日 | 2003-04-11 | | 公開日 | 2003-07-15 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II.
Mol.Cell, 11, 2003
|
|
1P1M
 
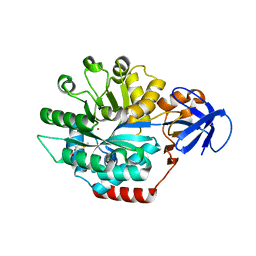 | | Structure of Thermotoga maritima amidohydrolase TM0936 bound to Ni and methionine | | 分子名称: | Hypothetical protein TM0936, METHIONINE, NICKEL (II) ION | | 著者 | Kniewel, R, Buglino, J.A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-04-12 | | 公開日 | 2003-04-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the hypothetical protein TM0936 from Thermotoga maritima at
1.5A bound to Ni and methionine
To be Published, 2003
|
|
1PSW
 
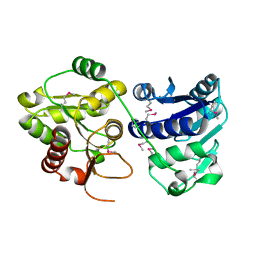 | | Structure of E. coli ADP-heptose lps heptosyltransferase II | | 分子名称: | ADP-HEPTOSE LPS HEPTOSYLTRANSFERASE II | | 著者 | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-06-21 | | 公開日 | 2003-07-08 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of E. coli ADP-heptose lps heptosyltransferase II
To be Published
|
|
1PUJ
 
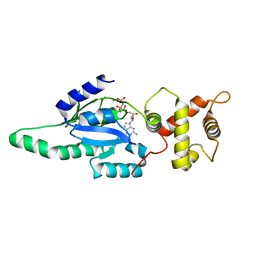 | | Structure of B. subtilis YlqF GTPase | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, conserved hypothetical protein ylqF | | 著者 | Kniewel, R, Buglino, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-06-24 | | 公開日 | 2003-07-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the YlqF GTPase from B. subtilis
To be Published
|
|
1PUG
 
 | | Structure of E. coli Ybab | | 分子名称: | Hypothetical UPF0133 protein ybaB | | 著者 | Kniewel, R, Buglino, J, Chadna, T, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-06-24 | | 公開日 | 2003-07-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of E. coli Ybab
To be Published
|
|
