6LMS
 
 | |
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | 登録日 | 2020-12-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | 分子名称: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
4RMK
 
 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in P65 crystal form | | 分子名称: | CALCIUM ION, Latrophilin-3 | | 著者 | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | 登録日 | 2014-10-21 | | 公開日 | 2015-08-19 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.606 Å) | | 主引用文献 | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
4RML
 
 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in C2221 crystal form | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Latrophilin-3, MAGNESIUM ION | | 著者 | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | 登録日 | 2014-10-21 | | 公開日 | 2015-08-19 | | 最終更新日 | 2015-10-07 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | 分子名称: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | 著者 | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | 登録日 | 2017-03-23 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | 著者 | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | 登録日 | 2020-04-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-22 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | 著者 | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | 登録日 | 2020-04-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-22 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | 著者 | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | 登録日 | 2020-04-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-22 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1HZ3
 
 | | ALZHEIMER'S DISEASE AMYLOID-BETA PEPTIDE (RESIDUES 10-35) | | 分子名称: | A-BETA AMYLOID | | 著者 | Zhang, S, Iwata, K, Lachenmann, M.J, Peng, J.W, Li, S, Stimson, E.R, Lu, Y, Felix, A.M, Maggio, J.E, Lee, J.P. | | 登録日 | 2001-01-23 | | 公開日 | 2001-01-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Alzheimer's peptide a beta adopts a collapsed coil structure in water.
J.Struct.Biol., 130, 2000
|
|
1PGV
 
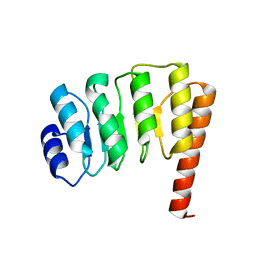 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | 分子名称: | tropomodulin TMD-1 | | 著者 | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2003-05-28 | | 公開日 | 2003-06-10 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
5V6I
 
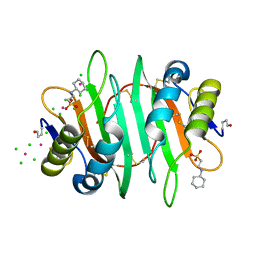 | | Glycan binding protein Y3 from mushroom Coprinus comatus possesses anti-leukemic activity - Pt derivative | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, PLATINUM (II) ION, ... | | 著者 | Li, K, Zhang, P, Gang, Y, Xia, C, Polston, J.E, Li, G, Li, S, Lin, Z, Yang, L.-J, Bruner, S.D, Ding, Y. | | 登録日 | 2017-03-16 | | 公開日 | 2017-08-16 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V6J
 
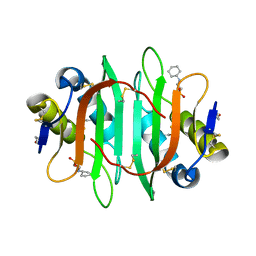 | | Glycan binding protein Y3 from mushroom Coprinus comatus possesses anti-leukemic activity | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TMV resistance protein Y3 | | 著者 | Li, K, Zhang, P, Gang, Y, Xia, C, Polston, J.E, Li, G, Li, S, Lin, Z, Yang, L.-J, Bruner, S.D, Ding, Y. | | 登録日 | 2017-03-16 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5J3J
 
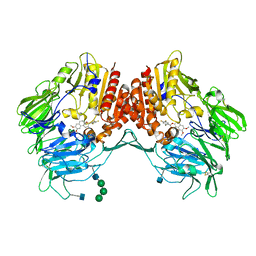 | | Crystal Structure of human DPP-IV in complex with HL1 | | 分子名称: | (2~{S},3~{R})-8,9-dimethoxy-3-[2,4,5-tris(fluoranyl)phenyl]-2,3-dihydro-1~{H}-benzo[f]chromen-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | 著者 | Wu, F, Li, H, Zhao, Z, Zhu, L, Xu, H, Li, S. | | 登録日 | 2016-03-31 | | 公開日 | 2017-04-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structure of human DPP-IV in complex with HL1
To Be Published
|
|
6TCL
 
 | | Photosystem I tetramer | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Chen, M, Perez-Boerema, A, Li, S, Amunts, A. | | 登録日 | 2019-11-06 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Distinct structural modulation of photosystem I and lipid environment stabilizes its tetrameric assembly.
Nat.Plants, 6, 2020
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | 分子名称: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | 著者 | Guo, Y, Wang, S, Nie, Y, Li, S. | | 登録日 | 2018-06-21 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
1J4I
 
 | | crystal structure analysis of the FKBP12 complexed with 000308 small molecule | | 分子名称: | 4-METHYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PENTANOIC ACID, FKBP12 | | 著者 | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | 登録日 | 2001-09-30 | | 公開日 | 2003-06-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
1J4H
 
 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | 分子名称: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | 著者 | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | 登録日 | 2001-09-30 | | 公開日 | 2003-06-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
6U9I
 
 | | Crystal structure of BvnE pinacolase from Penicillium brevicompactum | | 分子名称: | BvnE, DI(HYDROXYETHYL)ETHER, GLYCEROL | | 著者 | Ye, Y, Du, L, Zhang, X, Newmister, S.A, McCauley, M, Alegre-Requena, J.V, Zhang, W, Mu, S, Minami, A, Fraley, A.E, Adrover-Castellano, M.L, Carney, N, Shende, V.V, Oikawa, H, Kato, H, Tsukamoto, S, Paton, R.S, Williams, R.M, Sherman, D.H, Li, S. | | 登録日 | 2019-09-09 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.777 Å) | | 主引用文献 | Fungal-derived brevianamide assembly by a stereoselective semipinacolase.
Nat Catal, 3, 2020
|
|
7Y53
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | 著者 | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | 登録日 | 2022-06-16 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | 著者 | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | 登録日 | 2022-06-16 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | 著者 | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | 登録日 | 2022-06-16 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (4.51 Å) | | 主引用文献 | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
