5JCN
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | ASCORBIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5SXM
 
 | | WDR5 in complex with MLL Win motif peptidomimetic | | Descriptor: | ACE-ALA-ARG-THR-GLU-VAL-TYR-NH2, WD repeat-containing protein 5 | | Authors: | Alicea-Velazquez, N.L, Shinsky, S.A, Cosgrove, M.S. | | Deposit date: | 2016-08-09 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeted Disruption of the Interaction between WD-40 Repeat Protein 5 (WDR5) and Mixed Lineage Leukemia (MLL)/SET1 Family Proteins Specifically Inhibits MLL1 and SETd1A Methyltransferase Complexes.
J.Biol.Chem., 291, 2016
|
|
5TRL
 
 | |
5TRM
 
 | |
4ZMS
 
 | | Structure of the full-length response regulator spr1814 in complex with a phosphate analogue and B3C | | Descriptor: | 5-amino-2,4,6-tribromobenzene-1,3-diyl dihydroperoxide, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Chi, Y.M, Park, A. | | Deposit date: | 2015-05-04 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the full-length response regulator spr1814 in complex with a phosphate analogue reveals a novel conformational plasticity of the linker region
Biochem.Biophys.Res.Commun., 473, 2016
|
|
4ZMR
 
 | |
7XDP
 
 | |
7XDM
 
 | | ChCODH2 A559W mutant in anaerobic condition | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Heo, Y.Y, Kim, S.M. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | O2-tolerant CO dehydrogenase via tunnel redesign for the removal of CO from industrial flue gas
Nat Catal, 5, 2022
|
|
7XDN
 
 | |
3B2W
 
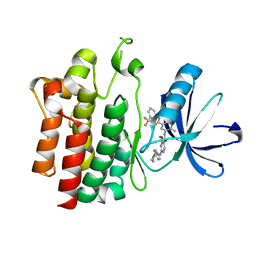 | | Crystal structure of pyrimidine amide 11 bound to Lck | | Descriptor: | N-[5-({[2-fluoro-3-(trifluoromethyl)phenyl]amino}carbonyl)-2-methylphenyl]-4-methoxy-2-[(4-piperazin-1-ylphenyl)amino]pyrimidine-5-carboxamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-(3-(phenylcarbamoyl)arylpyrimidine)-5-carboxamides as potent and selective inhibitors of Lck: structure, synthesis and SAR.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BMO
 
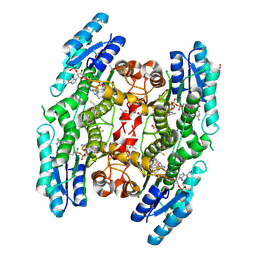 | | Structure of Pteridine Reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor (Compound AX4) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-[(4-methylphenyl)sulfanyl]pyrimidine-2,4-diamine, ... | | Authors: | Martini, V.P, Iulek, J, Hunter, W.N, Tulloch, L.B. | | Deposit date: | 2007-12-13 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3BYU
 
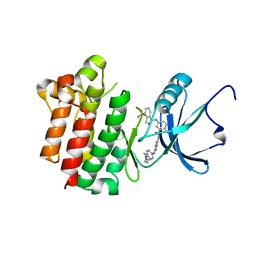 | | co-crystal structure of Lck and aminopyrimidine reverse amide 23 | | Descriptor: | 2-methyl-N-{4-methyl-3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-5-yl)carbamoyl]phenyl}-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of aminopyrimidine amides as potent, selective inhibitors of lymphocyte specific kinase: synthesis, structure-activity relationships, and inhibition of in vivo T cell activation.
J.Med.Chem., 51, 2008
|
|
3BMN
 
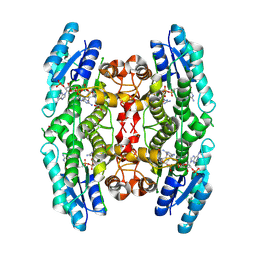 | | Structure of Pteridine Reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor (Compound AX3) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Martini, V.P, Iulek, J, Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2007-12-13 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3BMQ
 
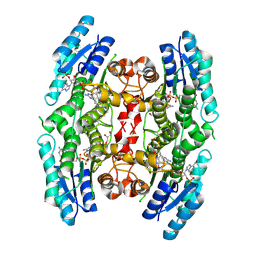 | | Structure of Pteridine Reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor (Compound AX5) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-(benzylsulfanyl)pyrimidine-2,4-diamine, ACETATE ION, ... | | Authors: | Martini, V.P, Iulek, J, Hunter, W.N. | | Deposit date: | 2007-12-13 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3BMC
 
 | |
3BYS
 
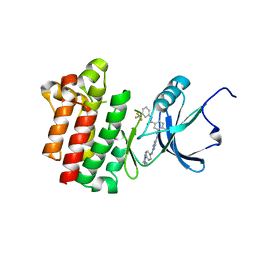 | | co-crystal structure of Lck and aminopyrimidine amide 10b | | Descriptor: | 4-methyl-N~3~-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-5-yl)-N~1~-[3-(trifluoromethyl)phenyl]benzene-1,3-dicarboxamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided design of aminopyrimidine amides as potent, selective inhibitors of lymphocyte specific kinase: synthesis, structure-activity relationships, and inhibition of in vivo T cell activation.
J.Med.Chem., 51, 2008
|
|
5X9U
 
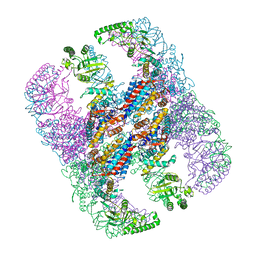 | | Crystal structure of group III chaperonin in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome, alpha subunit | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
5X9V
 
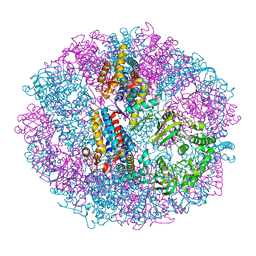 | | Crystal structure of group III chaperonin in the Closed state | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome, ... | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
3FLD
 
 | |
2OZ2
 
 | |
3JQC
 
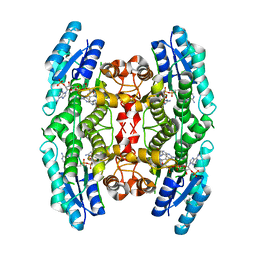 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-6-bromo-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile (JU2) | | Descriptor: | 2-amino-6-bromo-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
2P7U
 
 | |
2QZW
 
 | |
4P9M
 
 | | Crystal structure of 8ANC195 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC195 Fab heavy chain, 8ANC195 light chain | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2014-04-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Antibody 8ANC195 Reveals a Site of Broad Vulnerability on the HIV-1 Envelope Spike.
Cell Rep, 7, 2014
|
|
4P9H
 
 | |
