3AW8
 
 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Okada, K, Tsunoda, S, Taka, H, Baba, S, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
3AX6
 
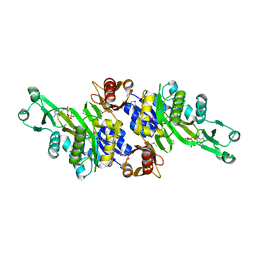 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoribosylaminoimidazole carboxylase, ATPase subunit | | Authors: | Miyazawa, R, Kanagawa, M, Baba, S, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
2OM6
 
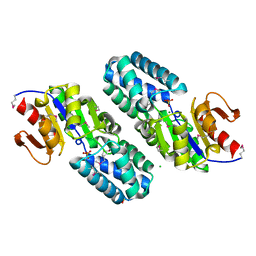 | | Hypothetical Protein (Probable Phosphoserine Phosph (PH0253) from Pyrococcus Horikoshii OT3 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable phosphoserine phosphatase, ... | | Authors: | Jeyakanthan, J, Vaijayanthimala, S, Gayathri, D, Velmurugan, D, Baba, S, Ebihara, A, Shinkai, A, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-21 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hypothetical Protein (Probable Phosphoserine Phosph (PH0253) from Pyrococcus Horikoshii OT3
To be Published
|
|
3CQ1
 
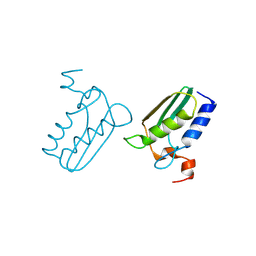 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (TT1362) from Thermus Thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB138 | | Authors: | Jeyakanthan, J, Satoh, S, Kitamura, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (TT1362) from Thermus Thermophilus HB8
To be Published
|
|
2PBQ
 
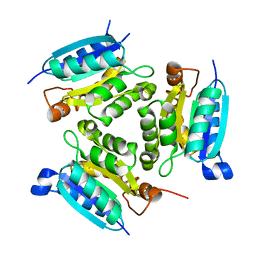 | | Crystal structure of molybdenum cofactor biosynthesis (aq_061) From aquifex aeolicus VF5 | | Descriptor: | Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Mahesh, S, Kanaujia, S.P, Ramakumar, S, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of molybdenum cofactor biosynthesis (aq_061) from aquifex aeolicus VF5
to be published
|
|
2QQ1
 
 | | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5 | | Descriptor: | Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Mahesh, S, Kanaujia, S.P, Ramakumar, S, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5
To be Published
|
|
3AAM
 
 | | Crystal structure of endonuclease IV from Thermus thermophilus HB8 | | Descriptor: | Endonuclease IV, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Asano, R, Ishikawa, H, Nakane, S, Baba, S, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-11-20 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | An additional C-terminal loop in endonuclease IV, an apurinic/apyrimidinic endonuclease, controls binding affinity to DNA
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ACC
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase with GMP from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3ACB
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1IZ9
 
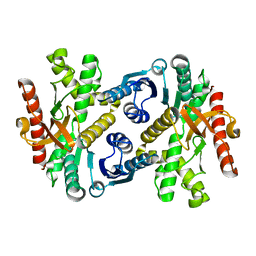 | | Crystal Structure of Malate Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | MALATE DEHYDROGENASE | | Authors: | Hirose, R, Hasegawa, T, Yamano, A, Kuramitsu, S, Hamada, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Malate Dehydrogenase from Thermus themrophilus HB8
To be published
|
|
3CQ2
 
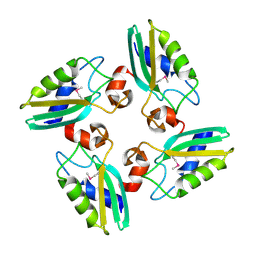 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (other form) from Thermus Thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB138 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Satoh, S, Kitamura, Y, Ebihara, A, Chen, L, Liu, Z.J, Wang, B.C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein from Thermus Thermophilus HB8
To be Published
|
|
3VUQ
 
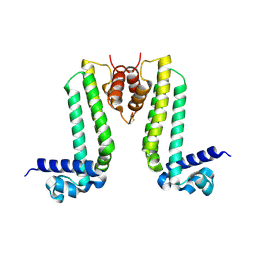 | | Crystal structure of TTHA0167, a transcriptional regulator, TetR/AcrR family from Thermus thermophilus HB8 | | Descriptor: | Transcriptional regulator (TetR/AcrR family) | | Authors: | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2012-07-04 | | Release date: | 2013-02-27 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and function of a TetR family transcriptional regulator, SbtR, from thermus thermophilus HB8
Proteins, 81, 2013
|
|
3VG8
 
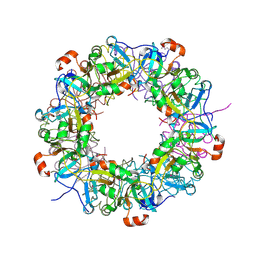 | | Crystal structure of hypothetical protein TTHB210 from Thermus thermophilus HB8 | | Descriptor: | Hypothetical Protein TTHB210 | | Authors: | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hypothetical protein TTHB210, controlled by the [sigma]E/anti-[sigma]E regulatory system in Thermus thermophilus HB8, reveals a novel homodecamer
Proteins, 80, 2012
|
|
3VPR
 
 | | Crystal Structure of a TetR Family Transcriptional Regulator PfmR from Thermus thermophilus HB8 | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Agari, Y, Sakamoto, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2012-03-12 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Transcriptional repression mediated by a TetR family protein, PfmR, from Thermus thermophilus HB8
J.Bacteriol., 2012
|
|
3W34
 
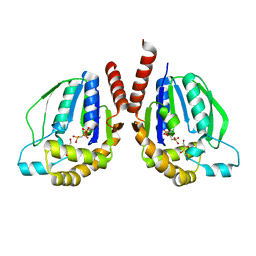 | | Ternary complex of Thermus thermophilus HB8 uridine-cytidine kinase with substrates | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Uridine kinase | | Authors: | Tomoike, F, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural and Biochemical Studies on the Reaction Mechanism of Uridine-Cytidine Kinase
Protein J., 34, 2015
|
|
3W8N
 
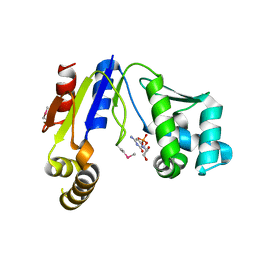 | |
3W5W
 
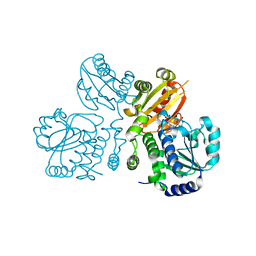 | | Mn2+-GMP complex of nanoRNase (Nrn) from Bacteroides fragilis | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Putative exopolyphosphatase-related protein | | Authors: | Uemura, Y, Nakagawa, N, Wakamatsu, T, Montelione, G.T, Hunt, J.F, Masui, R, Kuramitsu, S. | | Deposit date: | 2013-02-07 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of the ligand-binding form of nanoRNase from Bacteroides fragilis, a member of the DHH/DHHA1 phosphoesterase family of proteins.
Febs Lett., 587, 2013
|
|
3W90
 
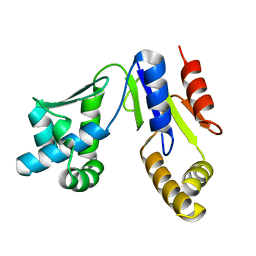 | |
3W8R
 
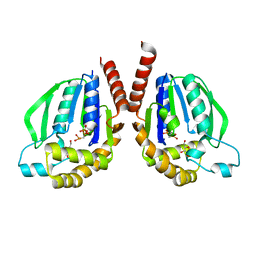 | | Mutant structure of Thermus thermophilus HB8 uridine-cytidine kinase | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Uridine kinase | | Authors: | Tomoike, F, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Indispensable residue for uridine binding in the uridine-cytidine kinase family.
Biochem Biophys Rep, 11, 2017
|
|
3AAL
 
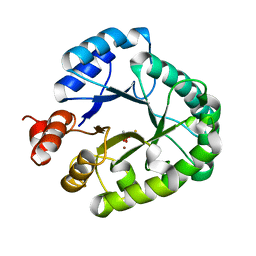 | | Crystal Structure of endonuclease IV from Geobacillus kaustophilus | | Descriptor: | CACODYLATE ION, FE (III) ION, Probable endonuclease 4, ... | | Authors: | Asano, R, Ishikawa, H, Nakane, S, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-11-20 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An additional C-terminal loop in endonuclease IV, an apurinic/apyrimidinic endonuclease, controls binding affinity to DNA
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3AU6
 
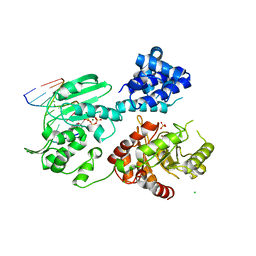 | | DNA polymerase X from Thermus thermophilus HB8 ternary complex with primer/template DNA and ddGTP | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*TP*(DDG))-3', 5'-D(*CP*GP*GP*CP*CP*AP*TP*AP*CP*TP*G)-3', ... | | Authors: | Nakane, S, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-28 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis of the kinetic mechanism of a gap-filling X-family DNA polymerase that binds Mg(2+)-dNTP before binding to DNA.
J.Mol.Biol., 417, 2012
|
|
3AU2
 
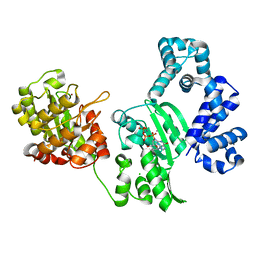 | | DNA polymerase X from Thermus thermophilus HB8 complexed with Ca-dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nakane, S, Ishikawa, H, Wakamatsu, T, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-28 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structural basis of the kinetic mechanism of a gap-filling X-family DNA polymerase that binds Mg(2+)-dNTP before binding to DNA.
J.Mol.Biol., 417, 2012
|
|
3AUO
 
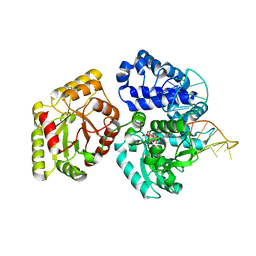 | | DNA polymerase X from Thermus thermophilus HB8 ternary complex with 1-nt gapped DNA and ddGTP | | Descriptor: | 1-nt gapped DNA, 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase beta family (X family), ... | | Authors: | Nakane, S, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-02-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of the kinetic mechanism of a gap-filling X-family DNA polymerase that binds Mg(2+)-dNTP before binding to DNA.
J.Mol.Biol., 417, 2012
|
|
3AYT
 
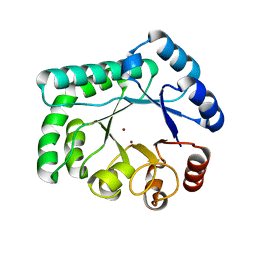 | | TTHB071 protein from Thermus thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB071, ZINC ION | | Authors: | Nakane, S, Wakamatsu, T, Fukui, K, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In vivo, in vitro, and x-ray crystallographic analyses suggest the involvement of an uncharacterized triose-phosphate isomerase (TIM) barrel protein in protection against oxidative stress
J.Biol.Chem., 286, 2011
|
|
3AYV
 
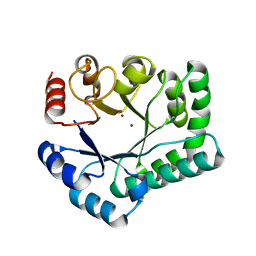 | | TTHB071 protein from Thermus thermophilus HB8 soaking with ZnCl2 | | Descriptor: | Putative uncharacterized protein TTHB071, ZINC ION | | Authors: | Nakane, S, Wakamatsu, T, Fukui, K, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In vivo, in vitro, and x-ray crystallographic analyses suggest the involvement of an uncharacterized triose-phosphate isomerase (TIM) barrel protein in protection against oxidative stress
J.Biol.Chem., 286, 2011
|
|
