1NWW
 
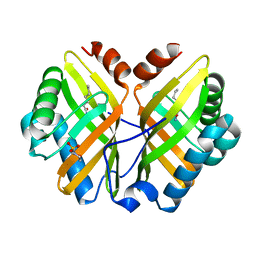 | | Limonene-1,2-epoxide hydrolase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEPTANAMIDE, Limonene-1,2-epoxide hydrolase | | Authors: | Arand, M, Hallberg, B.M, Zou, J, Bergfors, T, Oesch, F, van der Werf, M.J, de Bont, J.A.M, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site
EMBO J., 22, 2003
|
|
1OA3
 
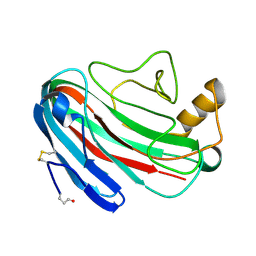 | | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1-4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Day, A.G, Jones, T.A, Mitchinson, C. | | Deposit date: | 2002-12-28 | | Release date: | 2003-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability
Protein Sci., 12, 2003
|
|
1OA2
 
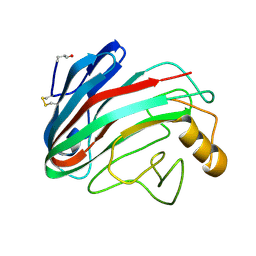 | | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Day, A.G, Jones, T.A, Mitchinson, C. | | Deposit date: | 2002-12-28 | | Release date: | 2003-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability
Protein Sci., 12, 2003
|
|
1NU3
 
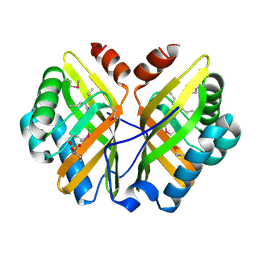 | | Limonene-1,2-epoxide hydrolase in complex with valpromide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PROPYLPENTANAMIDE, limonene-1,2-epoxide hydrolase | | Authors: | Arand, M, Hallberg, B.M, Zou, J, Bergfors, T, Oesch, F, van der Werf, M.J, de Bont, J.A.M, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site
EMBO J., 22, 2003
|
|
1OA4
 
 | | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability | | Descriptor: | ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Day, A.G, Jones, T.A, Mitchinson, C. | | Deposit date: | 2002-12-28 | | Release date: | 2003-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparison of Family 12 Glycoside Hydrolases and Recruited Substitutions Important for Thermal Stability
Protein Sci., 12, 2003
|
|
1OGM
 
 | |
1OGO
 
 | | Dex49A from Penicillium minioluteum complex with isomaltose | | Descriptor: | DEXTRANASE, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Larsson, A.M, Stahlberg, J, Jones, T.A. | | Deposit date: | 2003-05-08 | | Release date: | 2003-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dextranase from Penicillium Minioluteum. Reaction Course, Crystal Structure, and Product Complex
Structure, 11, 2003
|
|
1OLQ
 
 | | The Trichoderma reesei cel12a P201C mutant, structure at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
1OLR
 
 | | The Humicola grisea Cel12A Enzyme Structure at 1.2 A Resolution | | Descriptor: | ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
1Q2B
 
 | | CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, EXOCELLOBIOHYDROLASE I | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D.
J.Mol.Biol., 333, 2003
|
|
1Q2E
 
 | | CELLOBIOHYDROLASE CEL7A WITH LOOP DELETION 245-252 AND BOUND NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EXOCELLOBIOHYDROLASE I, ... | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D
J.Mol.Biol., 333, 2003
|
|
1PMP
 
 | |
1QO7
 
 | | Structure of Aspergillus niger epoxide hydrolase | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Zou, J.-Y, Hallberg, B.M, Bergfors, T, Oesch, F, Arand, M, Mowbray, S.L, Jones, T.A. | | Deposit date: | 1999-11-04 | | Release date: | 2000-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Aspergillus Niger Epoxide Hydrolase at 1.8A Resolution: Implications for the Structure and Function of the Mammalian Microsomal Class of Epoxide Hydrolases
Structure, 8, 2000
|
|
1RBL
 
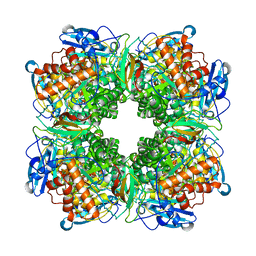 | | STRUCTURE DETERMINATION AND REFINEMENT OF RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM SYNECHOCOCCUS PCC6301 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Newman, J, Gutteridge, S, Branden, C.-I, Jones, T.A. | | Deposit date: | 1993-05-12 | | Release date: | 1994-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure determination and refinement of ribulose 1,5-bisphosphate carboxylase/oxygenase from Synechococcus PCC6301.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1RPJ
 
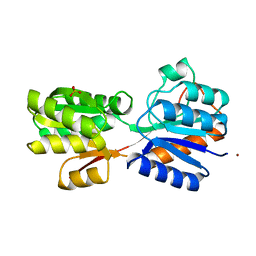 | | CRYSTAL STRUCTURE OF D-ALLOSE BINDING PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (PRECURSOR OF PERIPLASMIC SUGAR RECEPTOR), SULFATE ION, ZINC ION, ... | | Authors: | Chaudhuri, B, Jones, T.A, Mowbray, S.L. | | Deposit date: | 1999-02-04 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of D-allose binding protein from Escherichia coli bound to D-allose at 1.8 A resolution.
J.Mol.Biol., 286, 1999
|
|
2X1M
 
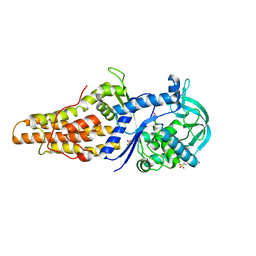 | | Crystal structure of Mycobacterium smegmatis methionyl-tRNA synthetase in complex with methionine | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DIHYDROGENPHOSPHATE ION, METHIONINE, ... | | Authors: | Ingvarsson, H, Jones, T.A, Unge, T. | | Deposit date: | 2009-12-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility and Communication within the Structure of the Mycobacterium Smegmatis Methionyl-tRNA Synthetase.
FEBS J., 277, 2010
|
|
2X1L
 
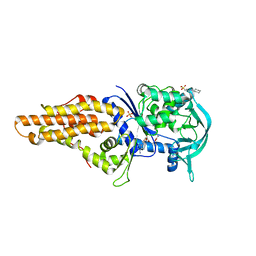 | | Crystal structure of Mycobacterium smegmatis methionyl-tRNA synthetase in complex with methionine and adenosine | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Ingvarsson, H, Jones, T.A, Unge, T. | | Deposit date: | 2009-12-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Flexibility and Communication within the Structure of the Mycobacterium Smegmatis Methionyl-tRNA Synthetase.
FEBS J., 277, 2010
|
|
1AXG
 
 | | CRYSTAL STRUCTURE OF THE VAL203->ALA MUTANT OF LIVER ALCOHOL DEHYDROGENASE COMPLEXED WITH COFACTOR NAD AND INHIBITOR TRIFLUOROETHANOL SOLVED TO 2.5 ANGSTROM RESOLUTION | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Colby, T.D, Chin, J.K, Bahnson, B.J, Goldstein, B.M, Klinman, J.P. | | Deposit date: | 1997-10-15 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A link between protein structure and enzyme catalyzed hydrogen tunneling.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6ADH
 
 | |
7ADH
 
 | |
1CBH
 
 | |
1ADC
 
 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLPICOLINAMIDE ADENINE-DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
1ADB
 
 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLNICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
1AXE
 
 | | CRYSTAL STRUCTURE OF THE ACTIVE-SITE MUTANT PHE93->TRP OF HORSE LIVER ALCOHOL DEHYDROGENASE IN COMPLEX WITH NAD AND INHIBITOR TRIFLUOROETHANOL | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Colby, T.D, Chin, J.K, Goldstein, B.M. | | Deposit date: | 1997-10-15 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A link between protein structure and enzyme catalyzed hydrogen tunneling.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
5CAC
 
 | | REFINED STRUCTURE OF HUMAN CARBONIC ANHYDRASE II AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CARBONIC ANHYDRASE FORM C, SULFITE ION, ZINC ION | | Authors: | Lindahl, M, Habash, D, Harrop, S, Helliwell, D.R, Liljas, A. | | Deposit date: | 1991-09-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of human carbonic anhydrase II at 2.0 A resolution.
Proteins, 4, 1988
|
|
