5CD3
 
 | | Structure of immature VRC01-class antibody DRVIA7 | | Descriptor: | DRVIA7 Heavy Chain, DRVIA7 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor.
Immunity, 44, 2016
|
|
5CD5
 
 | |
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp14/15 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp15/16 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp4/5 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp9/10 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp6/7 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E5X
 
 | |
8JTN
 
 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 2-propyl-2-azoniatricyclo[7.3.0.0^{3,7}]dodeca-1(9),2,7-trien-8-amine, Tudor domain-containing protein 3 | | Authors: | Chen, M, Wang, Z, Li, W, Shang, X, Liu, Y. | | Deposit date: | 2023-06-22 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Tudor domain of TDRD3 in complex with a small molecule antagonist.
Biochim Biophys Acta Gene Regul Mech, 1866, 2023
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7CKF
 
 | | The N-terminus of interferon-inducible antiviral protein-dimer | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | Authors: | Cui, W, Yang, H.T. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7W2P
 
 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 2-[(4-fluorophenyl)methyl]-2-azatricyclo[7.3.0.0^{3,7}]dodeca-1(9),3(7)-dien-8-imine, MAGNESIUM ION, Survival motor neuron protein, ... | | Authors: | Li, W, Arrowsmith, C.H, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
7W30
 
 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 1,2-dimethylquinolin-4-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Li, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
2HHE
 
 | | OXYGEN AFFINITY MODULATION BY THE N-TERMINI OF THE BETA CHAINS IN HUMAN AND BOVINE HEMOGLOBIN | | Descriptor: | HEMOGLOBIN (DEOXY) (ALPHA CHAIN), HEMOGLOBIN (DEOXY) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gilliland, G.L, Pechik, I, Fronticelli, C, Ji, X. | | Deposit date: | 1994-09-29 | | Release date: | 1995-01-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chloride ion independence of the Bohr effect in a mutant human hemoglobin beta (V1M+H2deleted).
J.Biol.Chem., 269, 1994
|
|
1GSD
 
 | |
1GSF
 
 | | GLUTATHIONE TRANSFERASE A1-1 COMPLEXED WITH ETHACRYNIC ACID | | Descriptor: | ETHACRYNIC ACID, GLUTATHIONE TRANSFERASE A1-1 | | Authors: | L'Hermite, G, Sinning, I, Cameron, A.D, Jones, T.A. | | Deposit date: | 1995-06-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of human alpha-class glutathione transferase A1-1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate.
Structure, 3, 1995
|
|
1GSE
 
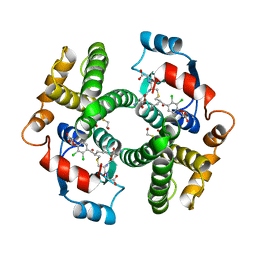 | |
8F4O
 
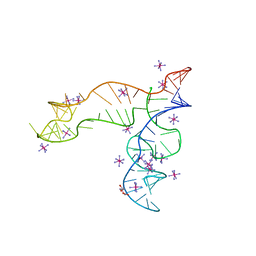 | | Apo structure of the TPP riboswitch aptamer domain | | Descriptor: | IRIDIUM HEXAMMINE ION, TETRAETHYLENE GLYCOL, TPP riboswitch aptamer domain, ... | | Authors: | Lee, H.-K, Wang, Y.-X, Stagno, J.R. | | Deposit date: | 2022-11-11 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Escherichia coli thiamine pyrophosphate-sensing riboswitch in the apo state.
Structure, 31, 2023
|
|
1M9B
 
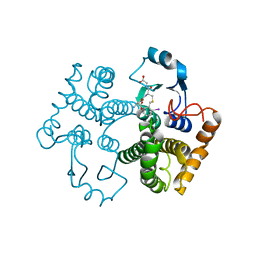 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with gamma-glutamyl[S-(2-iodobenzyl)cysteinyl]glycine | | Descriptor: | GAMMA-GLUTAMYL[S-(2-IODOBENZYL)CYSTEINYL]GLYCINE, Glutathione S-Transferase 26 kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M99
 
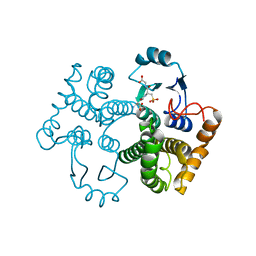 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with glutathione sulfonic acid | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-Transferase 26kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M9A
 
 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with S-hexylglutathione | | Descriptor: | Glutathione S-Transferase 26 kDa, S-HEXYLGLUTATHIONE | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
6LE0
 
 | |
8QPH
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra 14 crystals | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-02 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | VMXm - sub-micron microfocus beamline for macromolecular crystallography at Diamond Light Source
To Be Published
|
|
8QQC
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra single crystal | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-04 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | VMXm - sub-micron microfocus beamline for macromolecular crystallography at Diamond Light Source
To Be Published
|
|
