7P8E
 
 | | Crystal structure of the Receiver domain of M. truncatula cytokinin receptor MtCRE1 | | Descriptor: | CALCIUM ION, Receiver domain of histidine kinase | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
8BI3
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200W (crystal M200W#1) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8BKF
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200T (crystal M200T#o) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-09 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.221 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8BQO
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant M200I | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoaspartyl peptidase subunit alpha, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8C23
 
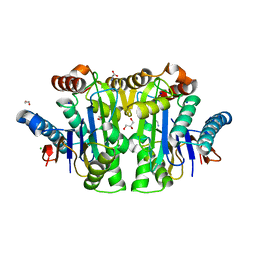 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200T (monoclinic form M200T#m) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-03 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
4Y31
 
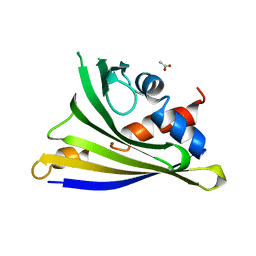 | | Crystal structure of yellow lupine LlPR-10.1A protein in ligand-free form | | Descriptor: | ACETATE ION, Protein LlR18A | | Authors: | Sliwiak, J, Michalska, K, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystallographic and CD probing of ligand-induced conformational changes in a plant PR-10 protein.
J.Struct.Biol., 193, 2016
|
|
3OFK
 
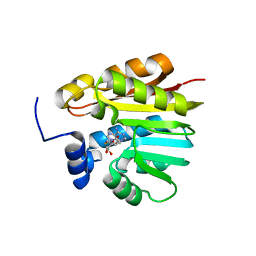 | | Crystal structure of N-methyltransferase NodS from Bradyrhizobium japonicum WM9 in complex with S-adenosyl-l-homocysteine (SAH) | | Descriptor: | Nodulation protein S, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cakici, O, Sikorski, M, Stepkowski, T, Bujacz, G, Jaskolski, M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of NodS N-Methyltransferase from Bradyrhizobium japonicum in Ligand-Free Form and as SAH Complex.
J.Mol.Biol., 404, 2010
|
|
3OFJ
 
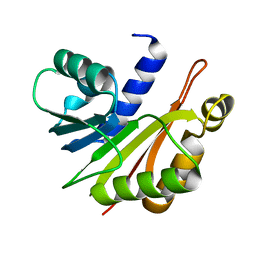 | | Crystal structure of N-methyltransferase NodS from Bradyrhizobium japonicum WM9 | | Descriptor: | Nodulation protein S | | Authors: | Cakici, O, Sikorski, M, Stepkowski, T, Bujacz, G, Jaskolski, M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of NodS N-Methyltransferase from Bradyrhizobium japonicum in Ligand-Free Form and as SAH Complex.
J.Mol.Biol., 404, 2010
|
|
7P8D
 
 | | Crystal structure of the Receiver domain of A. thaliana cytokinin receptor AtCRE1 in complex with Mg2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
7P8C
 
 | | Crystal structure of the Receiver domain of A. thaliana cytokinin receptor AtCRE1 in complex with K+ | | Descriptor: | POTASSIUM ION, Receiver domain of histidine kinase 4 | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
7Z0R
 
 | | AtWRKY18 DNA-binding domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, NITRATE ION, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
7Z0U
 
 | | Crystal structure of AtWRKY18 DNA-binding domain in complex with W-box DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*GP*GP*CP*G)-3'), WRKY transcription factor 18, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
8C0I
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200L (acyl-enzyme intermediate) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-12-16 | | Release date: | 2023-05-03 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8BP9
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200W (crystal M200W#2) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
1IHD
 
 | |
1JAZ
 
 | |
1JJA
 
 | |
1KHQ
 
 | | ORTHORHOMBIC FORM OF PAPAIN/ZLFG-DAM COVALENT COMPLEX | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
1KHP
 
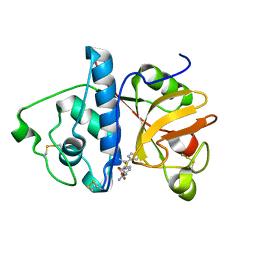 | | Monoclinic form of papain/ZLFG-DAM covalent complex | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
4LVC
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine | | Descriptor: | ACETATE ION, ADENOSINE, AMMONIUM ION, ... | | Authors: | Manszewski, T, Singh, K, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An enzyme captured in two conformational states: crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2HLH
 
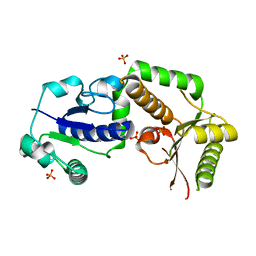 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | Nodulation fucosyltransferase, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
2HHC
 
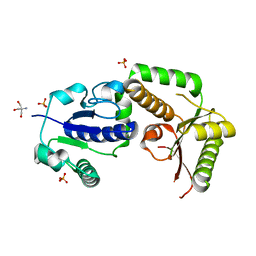 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
3US6
 
 | | Crystal Structure of Histidine-containing Phosphotransfer Protein MtHPt1 from Medicago truncatula | | Descriptor: | Histidine-containing Phosphotransfer Protein type 1, MtHPt1 | | Authors: | Ruszkowski, M, Brzezinski, K, Jedrzejczak, R, Dauter, M, Dauter, Z, Sikorski, M, Jaskolski, M. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Medicago truncatula histidine-containing phosphotransfer protein: Structural and biochemical insights into the cytokinin transduction pathway in plants.
Febs J., 280, 2013
|
|
5C9Y
 
 | |
2RCK
 
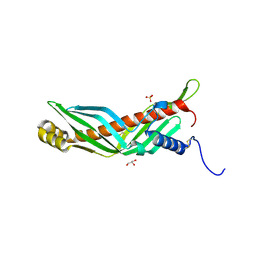 | | Crystal structure of juvenile hormone binding protein from Galleria mellonella hemolymph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Juvenile hormone binding protein, ... | | Authors: | Kolodziejczyk, R, Bujacz, G, Jakob, M, Ozyhar, A, Jaskolski, M, Kochman, M. | | Deposit date: | 2007-09-20 | | Release date: | 2008-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Insect Juvenile Hormone Binding Protein Shows Ancestral Fold Present in Human Lipid-Binding Proteins.
J.Mol.Biol., 377, 2008
|
|
