2TLX
 
 | | THERMOLYSIN (NATIVE) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1999-01-28 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
2VCI
 
 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-[2,4-DIHYDROXY-5-(1-METHYLETHYL)PHENYL]-N-ETHYL-4-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]ISOXAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
2V4C
 
 | |
2VCJ
 
 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-(5-chloro-2,4-dihydroxyphenyl)-N-ethyl-4-[4-(morpholin-4-ylmethyl)phenyl]isoxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
2XA5
 
 | |
2WX9
 
 | |
2WYP
 
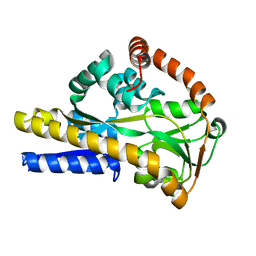 | | Crystal structure of sialic acid binding protein | | Descriptor: | SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP, deamino-beta-neuraminic acid | | Authors: | Fischer, M, Hubbard, R.E. | | Deposit date: | 2009-11-18 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water networks can determine the affinity of ligand binding to proteins.
J.Am.Chem.Soc., 2019
|
|
2WYK
 
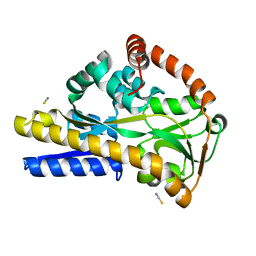 | | SiaP in complex with Neu5Gc | | Descriptor: | N-glycolyl-beta-neuraminic acid, SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP, THIOCYANATE ION | | Authors: | Fischer, M, Hubbard, R.E. | | Deposit date: | 2009-11-16 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water networks can determine the affinity of ligand binding to proteins.
J.Am.Chem.Soc., 2019
|
|
2WI2
 
 | | Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone | | Descriptor: | 4-METHYL-6-(METHYLSULFANYL)-1,3,5-TRIAZIN-2-AMINE, DIMETHYL SULFOXIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Barril, X, Borgognoni, J, Chene, P, Davies, N.G.M, Davis, B, Drysdale, M.J, Dymock, B, Eccles, S.A, Garcia-Echeverria, C, Fromont, C, Hayes, A, Hubbard, R.E, Jordan, A.M, Rugaard-Jensen, M, Massey, A, Merret, A, Padfield, A, Parsons, R, Radimerski, T, Raynaud, F.I, Robertson, A, Roughley, S.D, Schoepfer, J, Simmonite, H, Surgenor, A, Valenti, M, Walls, S, Webb, P, Wood, M, Workman, P, Wright, L.M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Combining Hit Identification Strategies: Fragment- Based and in Silico Approaches to Orally Active 2-Aminothieno[2,3-D]Pyrimidine Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 52, 2009
|
|
4INS
 
 | | THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | INSULIN (CHAIN A), INSULIN (CHAIN B), ZINC ION | | Authors: | Dodson, G.G, Dodson, E.J, Hodgkin, D.C, Isaacs, N.W, Vijayan, M. | | Deposit date: | 1989-07-10 | | Release date: | 1990-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of 2Zn pig insulin crystals at 1.5 A resolution.
Philos.Trans.R.Soc.London,Ser.B, 319, 1988
|
|
1ERR
 
 | |
1ERE
 
 | |
8HVA
 
 | |
8HV1
 
 | | Crystal structure of EGFR_DMX in complex with covalently bound fragment 1 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-(3-bromophenyl)-1,4-dihydro-1,2,4-triazole-5-thione, Epidermal growth factor receptor, ... | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
8HV4
 
 | | Crystal structure of EGFR_TMX in complex with covalently bound fragment 4 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
8HV5
 
 | | Crystal structure of EGFR_DMX in complex with compound 7 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Epidermal growth factor receptor, ~{N}-(1-methylbenzimidazol-4-yl)prop-2-enamide | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
8HV3
 
 | | Crystal structure of EGFR_DMX in complex with covalently bound fragment 4 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DIMETHYL SULFOXIDE, Epidermal growth factor receptor, ... | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
8HV6
 
 | | Crystal structure of EGFR_TMX in complex with covalently bound fragment 8 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DIMETHYL SULFOXIDE, Epidermal growth factor receptor, ... | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
8HV2
 
 | |
8HV8
 
 | | Crystal structure of EGFR_TMX in complex with covalently bound fragment 10 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Epidermal growth factor receptor, ~{N}-pyrazolo[1,5-a]pyridin-2-ylprop-2-enamide | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
8HV9
 
 | | Crystal structure of EGFR_TMX in complex with covalently bound fragment 12 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DIMETHYL SULFOXIDE, Epidermal growth factor receptor, ... | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
8HV7
 
 | | Crystal structure of EGFR_TMX in complex with covalently bound fragment 9 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DIMETHYL SULFOXIDE, Epidermal growth factor receptor, ... | | Authors: | Dokurno, P. | | Deposit date: | 2022-12-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A covalent fragment-based strategy targeting a novel cysteine to inhibit activity of mutant EGFR kinase.
Rsc Med Chem, 14, 2023
|
|
7NB4
 
 | | Structure of Mcl-1 complex with compound 1 | | Descriptor: | (2~{R})-2-[[5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
7NB7
 
 | | Structure of Mcl-1 complex with compound 6b | | Descriptor: | (2~{R})-2-[[7-but-2-ynyl-5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-pyrrolo[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
8CE3
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with 3D fragment 2548 | | Descriptor: | (3~{S})-1-ethanoyl-3-(4-methylphenyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Gilio, A.K, Darby, J.F, Wu, L, Davies, G.J. | | Deposit date: | 2023-02-01 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Modular 3-D Fragment Collection: Design, Synthesis and Screening
To Be Published
|
|
