7DRZ
 
 | |
7DRY
 
 | | Crystal structure of Aspergillus oryzae Rib2 deaminase | | Descriptor: | CMP/dCMP-type deaminase domain-containing protein, SULFATE ION, ZINC ION | | Authors: | Chen, S.C, Liaw, S.H, Hsu, C.H. | | Deposit date: | 2020-12-30 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of Aspergillus oryzae Rib2 deaminase: the functional mechanism involved in riboflavin biosynthesis.
Iucrj, 8, 2021
|
|
7VE6
 
 | | N-terminal domain of VraR | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator protein VraR | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7VE4
 
 | | C-terminal domain of VraR | | Descriptor: | DNA-binding response regulator | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7VE5
 
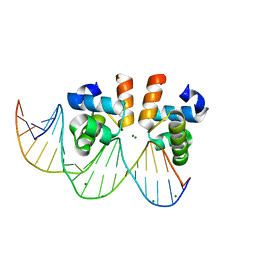 | | C-terminal domain of VraR | | Descriptor: | DNA-binding response regulator, MAGNESIUM ION, R1-DNA | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
5YB9
 
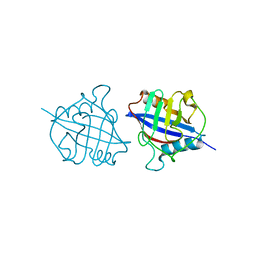 | | Crystal structure of a dimeric cyclophilin A from T.vaginalis | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Chou, C.C, Martin, T, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
5YBA
 
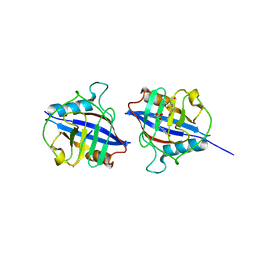 | | Dimeric Cyclophilin from T.vaginalis in complex with Myb1 peptide | | Descriptor: | Myb1 peptide, Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Martin, T, Chou, C.C, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
5ZDE
 
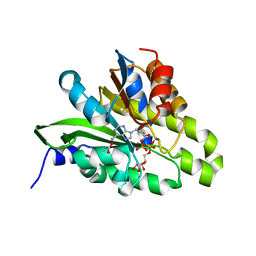 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P3221) | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDA
 
 | |
5ZDC
 
 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P32) | | Descriptor: | PHOSPHATE ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, poly ADP-ribose glycohydrolase | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDB
 
 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P21) | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDD
 
 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P212121) | | Descriptor: | PHOSPHATE ION, Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDF
 
 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) T267K mutant from Deinococcus radiodurans in complex with ADP-ribose | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDG
 
 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) T267R mutant from Deinococcus radiodurans in complex with ADP-ribose | | Descriptor: | Poly APD-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
6IDO
 
 | | Crystal structure of Klebsiella pneumoniae sigma4 of sigmaS fusing with the RNA polymerase beta-flap-tip-helix in complex with -35 element DNA | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*AP*TP*TP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*C)-3'), RNA polymerase sigma factor RpoS,RNA polymerase beta-flap-tip-helix | | Authors: | Lou, Y.C, Chien, C.Y, Chen, C, Hsu, C.H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.748 Å) | | Cite: | Structural basis for -35 element recognition by sigma4chimera proteins and their interactions with PmrA response regulator.
Proteins, 88, 2020
|
|
6JK2
 
 | | Crystal structure of a mini fungal lectin, PhoSL | | Descriptor: | Lectin, SULFATE ION | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
6JK3
 
 | | Crystal structure of a mini fungal lectin, PhoSL in complex with core-fucosylated chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lectin | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
6LFU
 
 | | Poa1p F152A mutant in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.123 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LCJ
 
 | |
6LFS
 
 | | Poa1p H23A mutant in complex with ADP-ribose | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFT
 
 | | Poa1p S30A mutant in complex with ADP-ribose | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFQ
 
 | | Crystal structure of Poa1p in apo form | | Descriptor: | ADP-ribose 1''-phosphate phosphatase, GLYCEROL | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LCL
 
 | |
6LFR
 
 | | Poa1p in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LCK
 
 | |
