4LPV
 
 | | Crystal structure of TENCON variant P41BR3-42 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TENCON variant P41BR3-42 | | Authors: | Teplyakov, A, Obmolova, G, Luo, J, Gilliland, G.L. | | Deposit date: | 2013-07-16 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-terminal beta-strand swapping in a consensus-derived fibronectin Type III scaffold.
Proteins, 82, 2014
|
|
4M6N
 
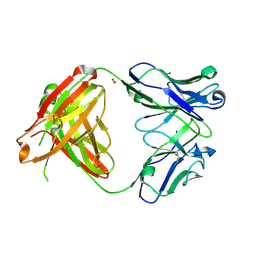 | |
4LPT
 
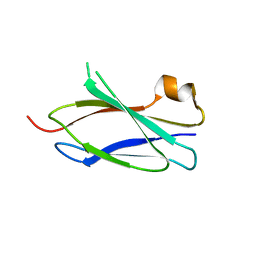 | |
4MAU
 
 | | Crystal structure of anti-ST2L antibody C2244 | | Descriptor: | C2244 heavy chain, C2244 light chain, FORMIC ACID, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4M6A
 
 | | N-Terminal beta-Strand Swapping in a Consensus Derived Alternative Scaffold Driven by Stabilizing Hydrophobic Interactions | | Descriptor: | Tencon | | Authors: | Luo, J, Teplyakov, A, Obmolova, G, Malia, T.J, Chan, W, Jocobs, S.A, O'neil, K.T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | N-terminal beta-strand swapping in a consensus-derived alternative scaffold driven by stabilizing hydrophobic interactions.
Proteins, 82, 2014
|
|
4M6O
 
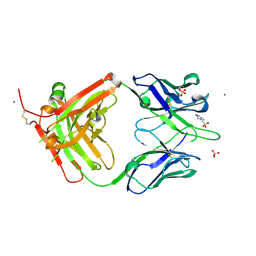 | | Crystal structure of anti-NGF antibody CNTO7309 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CNTO7309 heavy chain, CNTO7309 light chain, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4LPX
 
 | |
4M7K
 
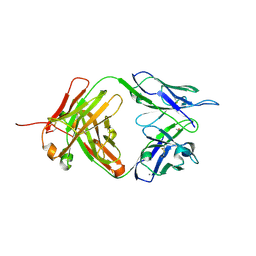 | | Crystal structure of anti-tissue factor antibody 10H10 | | Descriptor: | 10H10 heavy chain, 10H10 light chain, ACETATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4MA3
 
 | | Crystal structure of anti-hinge rabbit antibody C2095 | | Descriptor: | ACETATE ION, C2095 heavy chain, C2095 light chain, ... | | Authors: | Malia, T, Teplyakov, A, Gilliland, G.L. | | Deposit date: | 2013-08-15 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and specificity of an antibody targeting a proteolytically cleaved IgG hinge.
Proteins, 82, 2014
|
|
4M6M
 
 | | Crystal structure of anti-IL-23 antibody CNTO1959 at pH 9.5 | | Descriptor: | CNTO1959 heavy chain, CNTO1959 light chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4O51
 
 | | Crystal structure of the QAA variant of anti-hinge rabbit antibody 2095-2 in complex with IDES hinge peptide | | Descriptor: | IDES hinge peptide, QAA-2095-2 heavy chain, QAA-2095-2 light chain | | Authors: | Malia, T.J, Luo, J, Teplyakov, A, Gilliland, G.L. | | Deposit date: | 2013-12-19 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structure and specificity of an antibody targeting a proteolytically cleaved IgG hinge.
Proteins, 82, 2014
|
|
4O4Y
 
 | | Crystal structure of the anti-hinge rabbit antibody 2095-2 in complex with IDES hinge peptide | | Descriptor: | 2095-2 heavy chain, 2095-2 light chain, GLYCEROL, ... | | Authors: | Malia, T.J, Teplyakov, A, Luo, J, Gilliland, G.L. | | Deposit date: | 2013-12-19 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and specificity of an antibody targeting a proteolytically cleaved IgG hinge.
Proteins, 82, 2014
|
|
1N2A
 
 | | Crystal Structure of a Bacterial Glutathione Transferase from Escherichia coli with Glutathione Sulfonate in the Active Site | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase | | Authors: | Rife, C.L, Parsons, J.F, Xiao, G, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2002-10-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved structural elements in glutathione transferase homologues encoded in the genome of Escherichia coli
Proteins, 53, 2003
|
|
1MTC
 
 | | GLUTATHIONE TRANSFERASE MUTANT Y115F | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, Glutathione S-transferase YB1 | | Authors: | Ladner, J.E, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Local protein dynamics and catalysis: detection of segmental motion associated with rate-limiting product release by a glutathione transferase
Biochemistry, 41, 2002
|
|
1NIJ
 
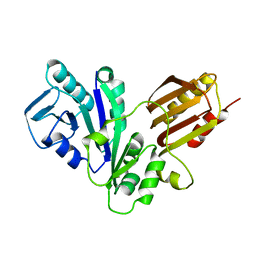 | | YJIA PROTEIN | | Descriptor: | Hypothetical protein yjiA | | Authors: | Khil, P.P, Obmolova, G, Teplyakov, A, Howard, A.J, Gilliland, G.L, Camerini-Otero, R.D, Structure 2 Function Project (S2F) | | Deposit date: | 2002-12-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function.
Proteins, 54, 2004
|
|
1NMO
 
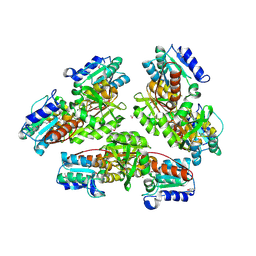 | | Structural genomics, protein ybgI, unknown function | | Descriptor: | FE (III) ION, Hypothetical protein ybgI | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1NMP
 
 | | Structural genomics, ybgI protein, unknown function | | Descriptor: | Hypothetical protein ybgI, MAGNESIUM ION | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1NRK
 
 | |
1PGA
 
 | |
1PGB
 
 | |
1R4B
 
 | |
1R45
 
 | | ADP-ribosyltransferase C3bot2 from Clostridium botulinum, triclinic form | | Descriptor: | GLYCEROL, Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L, Narumiya, S. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of ADP-ribosyltransferase C3bot2 from Clostridium botulinum
To be Published
|
|
1R4W
 
 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
1RW1
 
 | | YFFB (PA3664) PROTEIN | | Descriptor: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | Authors: | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-15 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
1S4C
 
 | | YHCH PROTEIN (HI0227) COPPER COMPLEX | | Descriptor: | ACETATE ION, COPPER (II) ION, Protein HI0227 | | Authors: | Teplyakov, A, Obmolova, G, Toedt, J, Gilliland, G.L. | | Deposit date: | 2004-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the bacterial YhcH protein indicates a role in sialic acid catabolism.
J.Bacteriol., 187, 2005
|
|
