1OCN
 
 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a cellobio-derived isofagomine at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, CALCIUM ION, ... | | Authors: | Varrot, A, Macdonald, J, Stick, R.V, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-02-09 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Distortion of a Cellobio-Derived Isofagomine Highlights the Potential Conformational Itinerary of Inverting Beta-Glucosidases
Chem.Commun.(Camb.), 21, 2003
|
|
1OHZ
 
 | | Cohesin-Dockerin complex from the cellulosome of Clostridium thermocellum | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A, ... | | Authors: | Carvalho, A.L, Dias, F.M.V, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Romao, M.J, Fontes, C.M.G.A. | | Deposit date: | 2003-06-04 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cellulosome Assembly Revealed by the Crystal Structure of the Cohesin-Dockerin Complex
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OH3
 
 | | E78R mutant of a carbohydrate binding module family 29 | | Descriptor: | NON-CATALYTIC PROTEIN 1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nurizzo, D, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-05-21 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand-Mediated Dimerization of a Carbohydrate -Binding Module Reveals a Novel Mechanism for Protein-Carbohydrate Recognition
J.Mol.Biol., 337, 2004
|
|
1ODT
 
 | | cephalosporin C deacetylase mutated, in complex with acetate | | Descriptor: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1QLD
 
 | |
2YG0
 
 | | CBM62 FROM CLOSTRIDIUM THERMOCELLUM XYL5A | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, Basle, A, Mckee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
2YFZ
 
 | | CBM62 FROM CLOSTRIDIUM THERMOCELLUM XYL5A | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, Basle, A, Mckee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
2Y8K
 
 | | Structure of CtGH5-CBM6, an arabinoxylan-specific xylanase. | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Firbank, S.J, Correia, M.A, Mazumder, K, Bras, J.L, Zhu, Y, Lewis, R.J, York, W.S, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure and Function of an Arabinoxylan-Specific Xylanase.
J.Biol.Chem., 286, 2011
|
|
2YB7
 
 | | CBM62 in complex with 6-alpha-D-Galactosyl-mannotriose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, McKee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-03-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
2XHJ
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. SeMet form of vCBM60. | | Descriptor: | CALCIUM ION, CALCIUM-DEPENDENT CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
2XHH
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
2XVL
 
 | | crystal structure of alpha-xylosidase (GH31) from Cellvibrio japonicus in complex with Pentaerythritol propoxylate (5 4 PO OH) | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, ALPHA-XYLOSIDASE, PUTATIVE, ... | | Authors: | Larsbrink, J, Izumi, A, Ibatullin, F, Nakhai, A, Gilbert, H.J, Davies, G.J, Brumer, H. | | Deposit date: | 2010-10-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Enzymatic Characterisation of a Glycoside Hydrolase Family 31 Alpha-Xylosidase from Cellvibrio Japonicus Involved in Xyloglucan Saccharification.
Biochem.J., 436, 2011
|
|
3W5M
 
 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Putative rhamnosidase | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
2XQO
 
 | | CtCel124: a cellulase from Clostridium thermocellum | | Descriptor: | Dockerin type 1, NICKEL (II) ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Carvalho, A.L, Verze, G, Bras, J.L.A, Cartmell, A, Bayer, E.A, Vazana, Y, Correia, M.A.S, Prates, J.A.M, Gilbert, H.J, Fontes, C.M.G.A, Romao, M.J. | | Deposit date: | 2010-09-06 | | Release date: | 2011-03-02 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights Into a Unique Cellulase Fold and Mechanism of Cellulose Hydrolysis
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-27 | | Release date: | 1999-07-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
2Y4J
 
 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH LACTATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LACTIC ACID, MANNOSYLGLYCERATE SYNTHASE, ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
2XVG
 
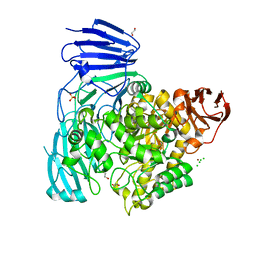 | | crystal structure of alpha-xylosidase (GH31) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, ALPHA XYLOSIDASE, CHLORIDE ION, ... | | Authors: | Larsbrink, J, Izumi, A, Ibatullin, F, Nakhai, A, Gilbert, H.J, Davies, G.J, Brumer, H. | | Deposit date: | 2010-10-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Enzymatic Characterisation of a Glycoside Hydrolase Family 31 Alpha-Xylosidase from Cellvibrio Japonicus Involved in Xyloglucan Saccharification.
Biochem.J., 436, 2011
|
|
2XFD
 
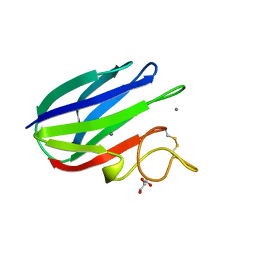 | | vCBM60 in complex with cellobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, GLYCEROL, ... | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2Y4L
 
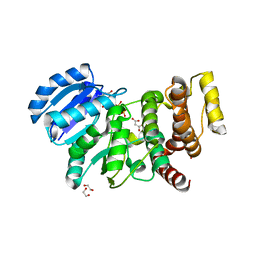 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH Manganese and GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, MALONATE ION, ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
2XVK
 
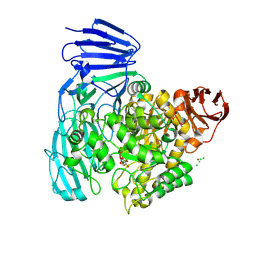 | | crystal structure of alpha-xylosidase (GH31) from Cellvibrio japonicus in complex with 5-fluoro-alpha-D-xylopyranosyl fluoride | | Descriptor: | (2R,3S,5R,6S)-2,6-DIFLUOROOXANE-3,4,5-TRIOL, ALPHA-XYLOSIDASE, PUTATIVE, ... | | Authors: | Larsbrink, J, Izumi, A, Ibatullin, F, Nakhai, A, Gilbert, H.J, Davies, G.J, Brumer, H. | | Deposit date: | 2010-10-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural and Enzymatic Characterisation of a Glycoside Hydrolase Family 31 Alpha-Xylosidase from Cellvibrio Japonicus Involved in Xyloglucan Saccharification.
Biochem.J., 436, 2011
|
|
2YFU
 
 | | CBM62 FROM CLOSTRIDIUM THERMOCELLUM XYL5A | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, Basle, A, Mckee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-04-08 | | Release date: | 2011-05-18 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
2YPJ
 
 | | Non-catalytic carbohydrate binding module CBM65B | | Descriptor: | ENDOGLUCANASE CEL5A, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Temple, M.J, Rogowski, A, Basle, A, Xue, J, Knox, J.P, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
2Y4M
 
 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH GDP-Mannose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
2Y4K
 
 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH MG-GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
3ZXJ
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
