7AXX
 
 | |
7AXP
 
 | |
7AXS
 
 | |
5FV6
 
 | | KpFlo11 presents a novel member of the Flo11 family with a unique recognition pattern for homophilic interactions | | Descriptor: | ACETATE ION, Flocculation protein FLO11, GLYCEROL, ... | | Authors: | Kraushaar, T, Brueckner, S, Mikolaiski, M, Schreiner, F, Veelders, M, Moesch, H.U, Essen, L.O. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family.
Elife, 9, 2020
|
|
5FV5
 
 | | KpFlo11 presents a novel member of the Flo11 family with a unique recognition pattern for homophilic interactions | | Descriptor: | ACETATE ION, Flocculation protein FLO11 | | Authors: | Kraushaar, T, Brueckner, S, Mikolaiski, M, Schreiner, F, Veelders, M, Moesch, H.U, Essen, L.O. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family.
Elife, 9, 2020
|
|
6CMV
 
 | | Crystal structure of Archaeal Biofilm Regulator (AbfR2) from Sulfolobus acidocaldarius | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Transcriptional regulator Lrs14-like protein | | Authors: | Essen, L.-O, Vogt, M.S, Banerjee, A. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of an Lrs14-like archaeal biofilm regulator from Sulfolobus acidocaldarius.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FN3
 
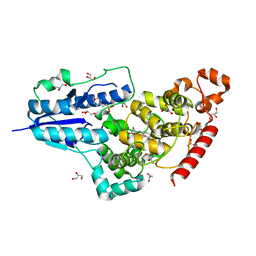 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
6FN0
 
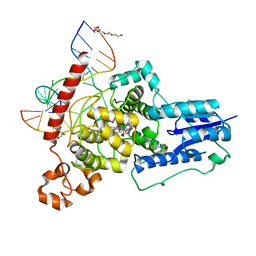 | | The animal-like Cryptochrome from Chlamydomonas reinhardtii in complex with 6-4 DNA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
6FN2
 
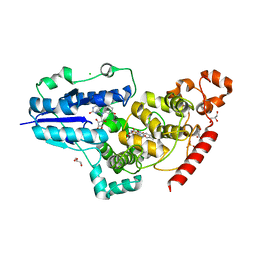 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
3CKY
 
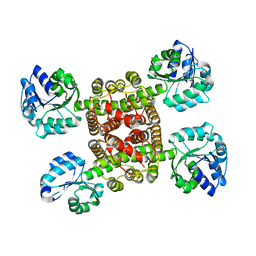 | |
1QAS
 
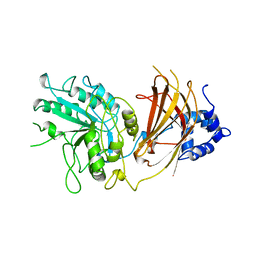 | |
1QAT
 
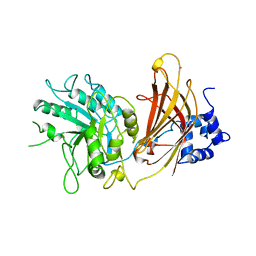 | |
7ZXV
 
 | | Orange Carotenoid Protein Trp-288 BTA mutant | | Descriptor: | CHLORIDE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one, ... | | Authors: | Moldenhauer, M, Tseng, H.-W, Kraskov, A, Tavraz, N.N, Hildebrandt, P, Hochberg, G, Essen, L.-O, Budisa, N, Korf, L, Maksimov, E.G, Friedrich, T. | | Deposit date: | 2022-05-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parameterization of a single H-bond in Orange Carotenoid Protein by atomic mutation reveals principles of evolutionary design of complex chemical photosystems.
Front Mol Biosci, 10, 2023
|
|
2VSQ
 
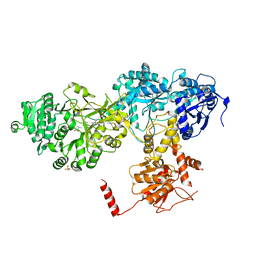 | | Structure of surfactin A synthetase C (SrfA-C), a nonribosomal peptide synthetase termination module | | Descriptor: | LEUCINE, SULFATE ION, SURFACTIN SYNTHETASE SUBUNIT 3 | | Authors: | Tanovic, A, Samel, S.A, Essen, L.-O, Marahiel, M.A. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the termination module of a nonribosomal peptide synthetase.
Science, 321, 2008
|
|
7PZA
 
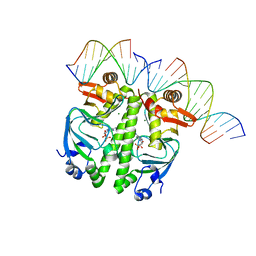 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
7PZB
 
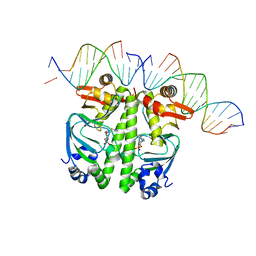 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
8A1H
 
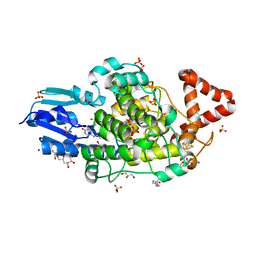 | | Bacterial 6-4 photolyase from Vibrio cholerase | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-4 photolyase (FeS-BCP, ... | | Authors: | Essen, L.-O, Emmerich, H.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Analysis of a Prokaryotic (6-4) Photolyase from the Aquatic Pathogen Vibrio Cholerae † .
Photochem.Photobiol., 99, 2023
|
|
1ASX
 
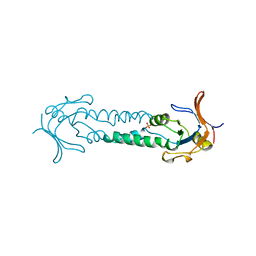 | | APICAL DOMAIN OF THE CHAPERONIN FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PHOSPHATE ION, THERMOSOME | | Authors: | Klumpp, M, Baumeister, W, Essen, L.-O. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Cell(Cambridge,Mass.), 91, 1997
|
|
1ASS
 
 | | APICAL DOMAIN OF THE CHAPERONIN FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PHOSPHATE ION, SODIUM ION, THERMOSOME | | Authors: | Klumpp, M, Baumeister, W, Essen, L.-O. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Cell(Cambridge,Mass.), 91, 1997
|
|
1FL2
 
 | |
2XC6
 
 | |
1FX2
 
 | |
6R1E
 
 | | Structure of dodecin from Streptomyces coelicolor | | Descriptor: | CHLORIDE ION, COENZYME A, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Essen, L.-O, Sander, B. | | Deposit date: | 2019-03-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparative biochemical and structural analysis of the flavin-binding dodecins fromStreptomyces davaonensisandStreptomyces coelicolorreveals striking differences with regard to multimerization.
Microbiology (Reading, Engl.), 165, 2019
|
|
6RY5
 
 | | Crystal structure of Dfg5 from Chaetomium thermophilum in complex with alpha-1,6-mannobiose | | Descriptor: | CALCIUM ION, Mannan endo-1,6-alpha-mannosidase, TRIETHYLENE GLYCOL, ... | | Authors: | Essen, L.-O, Vogt, M.S. | | Deposit date: | 2019-06-10 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural base for the transfer of GPI-anchored glycoproteins into fungal cell walls.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RY6
 
 | |
