3JUU
 
 | | Crystal structure of porphyranase B (PorB) from Zobellia galactanivorans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Hehemann, J.H, Correc, G, Barbeyron, T, Michel, G, Czjzek, M. | | Deposit date: | 2009-09-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota.
Nature, 464, 2010
|
|
5OLC
 
 | |
3P2N
 
 | | Discovery and structural characterization of a new glycoside hydrolase family abundant in coastal waters that was annotated as 'hypothetical protein' | | Descriptor: | 3,6-anhydro-alpha-L-galactosidase, CHLORIDE ION, ZINC ION | | Authors: | Rebuffet, E, Barbeyron, T, Czjzek, M, Michel, G. | | Deposit date: | 2010-10-03 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and structural characterization of a novel glycosidase family of marine origin.
Environ Microbiol, 13, 2011
|
|
5OPQ
 
 | | A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-D-galactosidase, ... | | Authors: | Ficko-Blean, E, Michel, G, Czjzek, M. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria.
Nat Commun, 8, 2017
|
|
1GX7
 
 | | Best model of the electron transfer complex between cytochrome c3 and [Fe]-hydrogenase | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Elantak, L, Morelli, X, Bornet, O, Hatchikian, C, Czjzek, M, Dolla, A, Guerlesquin, F. | | Deposit date: | 2002-03-28 | | Release date: | 2003-07-31 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | The Cytochrome C(3)-[Fe]-Hydrogenase Electron-Transfer Complex: Structural Model by NMR Restrained Docking
FEBS Lett., 548, 2003
|
|
2UWC
 
 | | Crystal structure of Nasturtium xyloglucan hydrolase isoform NXG2 | | Descriptor: | CELLULASE | | Authors: | Baumann, M.J, Eklof, J.M, Michel, G, Kallas, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
2UWA
 
 | | Crystal structure of the Nasturtium seedling xyloglucanase isoform NXG1 | | Descriptor: | CELLULASE, GLYCEROL | | Authors: | Baumann, M.J, Eklof, J, Michel, G, Kallasa, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-19 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
2UWB
 
 | | Crystal structure of the Nasturtium seedling mutant xyloglucanase isoform NXG1-delta-YNIIG | | Descriptor: | CELLULASE | | Authors: | Baumann, M.J, Eklof, J, Michel, G, Kallasa, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
1DK0
 
 | | CRYSTAL STRUCTURE OF THE HEMOPHORE HASA FROM SERRATIA MARCESCENS CRYSTAL FORM P2(1), PH8 | | Descriptor: | HEME-BINDING PROTEIN A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnoux, P, Haser, R, Izadi-Pruneyre, N, Lecroisey, A, Czjzek, M. | | Deposit date: | 1999-12-06 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms.
Proteins, 41, 2000
|
|
1DKH
 
 | | CRYSTAL STRUCTURE OF THE HEMOPHORE HASA, PH 6.5 | | Descriptor: | HEME-BINDING PROTEIN A, PROTOPORPHYRIN IX CONTAINING FE, SAMARIUM (III) ION, ... | | Authors: | Arnoux, P, Haser, R, Izadi-Pruneyre, N, Lecroisey, A, Czjzek, M. | | Deposit date: | 1999-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms.
Proteins, 41, 2000
|
|
1CEX
 
 | | STRUCTURE OF CUTINASE | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Czjzek, M, Lamzin, V, Nicolas, A, Cambillau, C. | | Deposit date: | 1997-02-18 | | Release date: | 1997-08-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution (1.0 A) crystal structure of Fusarium solani cutinase: stereochemical analysis.
J.Mol.Biol., 268, 1997
|
|
7QNM
 
 | | Crystallization and structural analyses of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION | | Authors: | Grigorian, E, Roret, T, Leblanc, C, Delage, L, Czjzek, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a l-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 32, 2023
|
|
1E08
 
 | | Structural model of the [Fe]-Hydrogenase/cytochrome c553 complex combining NMR and soft-docking | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Morelli, X, Czjzek, M, Hatchikian, C.E, Bornet, O, Fontecilla-Camps, J.C, Palma, N.P, Moura, J.J.G, Guerlesquin, F. | | Deposit date: | 2000-03-13 | | Release date: | 2000-08-25 | | Last modified: | 2019-11-27 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Fe-Hydrogenase/Cytochrome C553 Complex Combining Transverse Relaxation-Optimized Spectroscopy Experiments and Soft Docking Calculations.
J.Biol.Chem., 275, 2000
|
|
4ATF
 
 | | Crystal structure of inactivated mutant beta-agarase B in complex with agaro-octaose | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE B, SODIUM ION | | Authors: | Bernard, T, Hehemann, J.H, Correc, G, Jam, M, Michel, G, Czjzek, M. | | Deposit date: | 2012-05-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Characterization of the Complex Agarolytic Enzyme System from the Marine Bacterium Zobellia Galactanivorans.
J.Biol.Chem., 287, 2012
|
|
4ATE
 
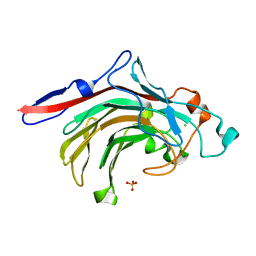 | | High resolution crystal structure of beta-porphyranase A from Zobellia galactanivorans | | Descriptor: | BETA-PORPHYRANASE A, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hehemann, J.H, Correc, G, Jam, M, Michel, G, Czjzek, M. | | Deposit date: | 2012-05-06 | | Release date: | 2012-07-25 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Biochemical and Structural Characterization of the Complex Agarolytic Enzyme System from the Marine Bacterium Zobellia Galactanivorans.
J.Biol.Chem., 287, 2012
|
|
7AJ0
 
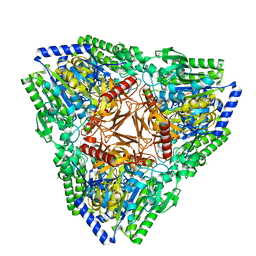 | | Crystal structure of PsFucS1 sulfatase from Pseudoalteromonas sp. | | Descriptor: | Arylsulfatase, CALCIUM ION, CHLORIDE ION | | Authors: | Roret, T, Mikkelsen, M.D, Czjzek, M, Meyer, A.S. | | Deposit date: | 2020-09-28 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A novel thermostable prokaryotic fucoidan active sulfatase PsFucS1 with an unusual quaternary hexameric structure.
Sci Rep, 11, 2021
|
|
7ARP
 
 | | Native L-2-haloacid dehalogenase from Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION, THIOCYANATE ION | | Authors: | Grigorian, E, Roret, T, Czjzek, M, Leblanc, C, Delage, L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 2022
|
|
7ASZ
 
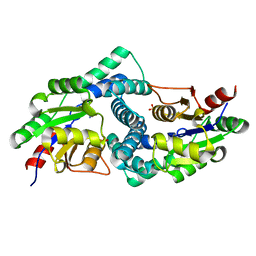 | | L-2-haloacid dehalogenase H190A mutant from Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION, THIOCYANATE ION | | Authors: | Grigorian, E, Roret, T, Czjzek, M, Leblanc, C, Delage, L. | | Deposit date: | 2020-10-28 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 2022
|
|
1EDG
 
 | |
2V5C
 
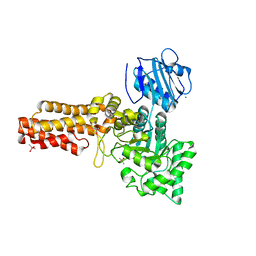 | | Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure | | Descriptor: | CACODYLATE ION, CALCIUM ION, O-GLCNACASE NAGJ, ... | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
2V5D
 
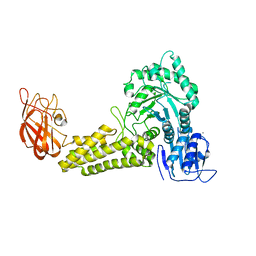 | | Structure of a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. | | Descriptor: | CALCIUM ION, O-GLCNACASE NAGJ | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
2W1N
 
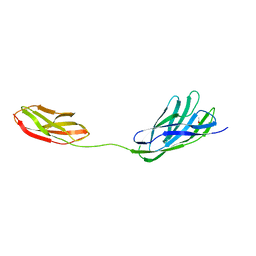 | | cohesin and fibronectin type-III double module construct from the Clostridium perfringens glycoside hydrolase GH84C | | Descriptor: | ACETATE ION, O-GLCNACASE NAGJ | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Czjzek, M, Smith, S.J, Boraston, A.B. | | Deposit date: | 2008-10-17 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
2YA1
 
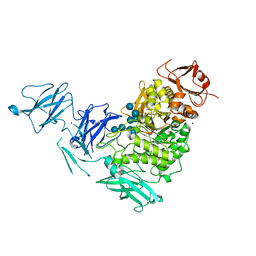 | | Product complex of a multi-modular glycogen-degrading pneumococcal virulence factor SpuA | | Descriptor: | PUTATIVE ALKALINE AMYLOPULLULANASE, SODIUM ION, SULFATE ION, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
2YA2
 
 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA in complex with an inhibitor. | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, PUTATIVE ALKALINE AMYLOPULLULANASE, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
2YA0
 
 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALKALINE AMYLOPULLULANASE, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
