2H6O
 
 | | Epstein Barr Virus Major Envelope Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Major outer envelope glycoprotein gp350, ... | | Authors: | Chen, X.S. | | Deposit date: | 2006-05-31 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Epstein-Barr virus major envelope glycoprotein
Nat.Struct.Mol.Biol., 13, 2006
|
|
5W1X
 
 | | Crystal Structure of Humanpapillomavirus18 (HPV18) Capsid L1 Pentamers Bound to Heparin Oligosaccharides | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, ... | | Authors: | Chen, X.S, Dasgupta, J. | | Deposit date: | 2017-06-05 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.374 Å) | | Cite: | Structural basis of oligosaccharide receptor recognition by human papillomavirus.
J. Biol. Chem., 286, 2011
|
|
2I0I
 
 | | X-ray crystal structure of Sap97 PDZ3 bound to the C-terminal peptide of HPV18 E6 | | Descriptor: | Disks large homolog 1, peptide E6 | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
2I04
 
 | | X-ray crystal structure of MAGI-1 PDZ1 bound to the C-terminal peptide of HPV18 E6 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, SULFATE ION, ... | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-09 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
2I0L
 
 | | X-ray crystal structure of Sap97 PDZ2 bound to the C-terminal peptide of HPV18 E6. | | Descriptor: | Disks large homolog 1, peptide E6 | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
1GHQ
 
 | | CR2-C3D COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, COMPLEMENT C3, CR2/CD121/C3D/EPSTEIN-BARR VIRUS RECEPTOR, ... | | Authors: | Szakonyi, G, Guthridge, J.M, Li, D, Holers, V.M, Chen, X.S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of complement receptor 2 in complex with its C3d ligand.
Science, 292, 2001
|
|
1LTL
 
 | | THE DODECAMER STRUCTURE OF MCM FROM ARCHAEAL M. THERMOAUTOTROPHICUM | | Descriptor: | DNA replication initiator (Cdc21/Cdc54), ZINC ION | | Authors: | Fletcher, R.J, Bishop, B.E, Leon, R.P, Sclafani, R.A, Ogata, C.M, Chen, X.S. | | Deposit date: | 2002-05-20 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure and function of MCM from archaeal M. Thermoautotrophicum
Nat.Struct.Biol., 10, 2003
|
|
1N25
 
 | | Crystal structure of the SV40 Large T antigen helicase domain | | Descriptor: | Large T Antigen, ZINC ION | | Authors: | Li, D, Zhao, R, Lilyestrom, W, Gai, D, Zhang, R, DeCaprio, J.A, Fanning, E, Jochimiak, A, Szakonyi, G, Chen, X.S. | | Deposit date: | 2002-10-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen
Nature, 423, 2003
|
|
2H1L
 
 | |
5K83
 
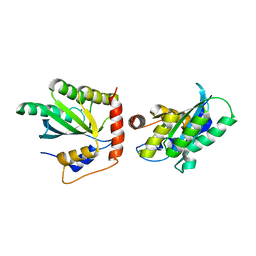 | | Crystal Structure of a Primate APOBEC3G N-Domain, in Complex with ssDNA | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
5K81
 
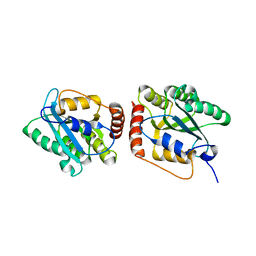 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
5K82
 
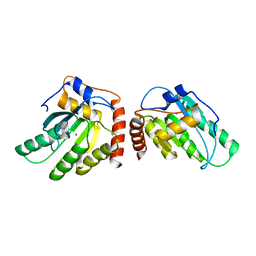 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
3IQS
 
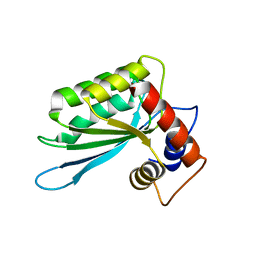 | | Crystal structure of the anti-viral APOBEC3G catalytic domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L.G, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
6X91
 
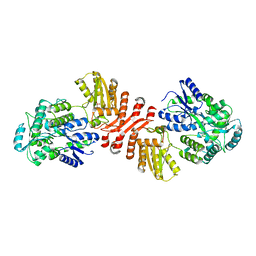 | | Crystal structure of MBP-fused human APOBEC1 | | Descriptor: | CACODYLATE ION, Maltodextrin-binding protein, C->U-editing enzyme APOBEC-1 chimera, ... | | Authors: | Wolfe, A.D, Li, S.-X, Chen, X.S. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The structure of APOBEC1 and insights into its RNA and DNA substrate selectivity.
NAR Cancer, 2, 2020
|
|
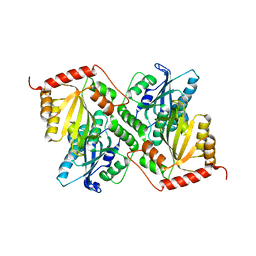
1SVL
 
 | | Co-crystal structure of SV40 large T antigen helicase domain and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
1SVM
 
 | | Co-crystal structure of SV40 large T antigen helicase domain and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
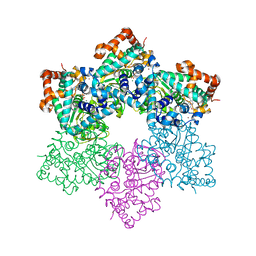
1SVO
 
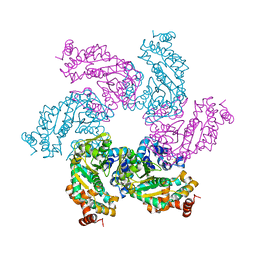 | | Structure of SV40 large T antigen helicase domain | | Descriptor: | ZINC ION, large T antigen | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
6P3X
 
 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.0) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
6P3Z
 
 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 5.2) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
6P3Y
 
 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.4) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
6P40
 
 | | Crystal Structure of Full Length APOBEC3G FKL | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
8UD3
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, consensus form | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8UD2
 
 | |
8UD4
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 1 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8UD5
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 2 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
