7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | 分子名称: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-20 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
6CZP
 
 | | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-04-09 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN.
To Be Published
|
|
7M1Y
 
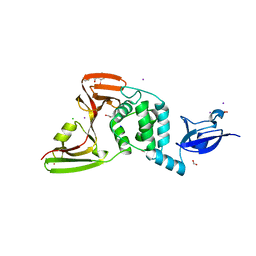 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with ebselen | | 分子名称: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Maltseva, N, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-03-15 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with ebselen
to be published
|
|
6DB1
 
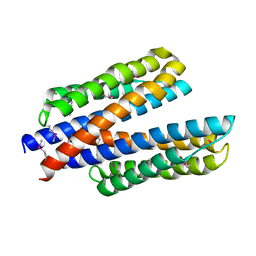 | | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica | | 分子名称: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-05-02 | | 公開日 | 2018-05-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.
To Be Published
|
|
6DFU
 
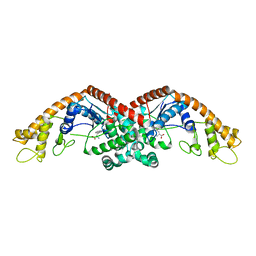 | | Tryptophan--tRNA ligase from Haemophilus influenzae. | | 分子名称: | TRYPTOPHAN, Tryptophan--tRNA ligase | | 著者 | Osipiuk, J, Maltseva, N, Mulligan, R, Grimshaw, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-05-15 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Tryptophan--tRNA ligase from Haemophilus influenzae.
to be published
|
|
6U13
 
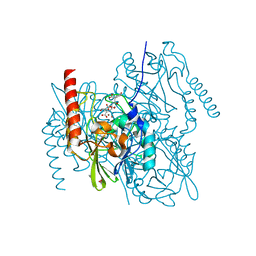 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed antibiotic moxalactam | | 分子名称: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-08-15 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed antibiotic moxalactam.
To Be Published
|
|
6U2Y
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions | | 分子名称: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, NICKEL (II) ION, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-08-20 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions
To Be Published
|
|
6UAC
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with cadmium and hydrolyzed moxolactam | | 分子名称: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, CADMIUM ION, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-09-10 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with cadmium and hydrolyzed moxolactam
To Be Published
|
|
6U2Z
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions | | 分子名称: | (2R)-2-[(1R)-1-{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3 -oxazine-4-carboxylic acid, 1,2-ETHANEDIOL, COPPER (II) ION, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-08-21 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions
To Be Published
|
|
6UAF
 
 | | Crystal Structure of the Metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Imipnem | | 分子名称: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-09-10 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of the Metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Imipnem
To Be Published
|
|
6U9C
 
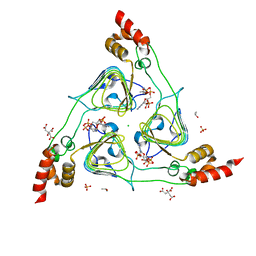 | | The 2.2 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the complex with Acetyl CoA | | 分子名称: | ACETYL COENZYME *A, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-09-07 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The 2.2 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the complex with Acetyl CoA
To Be Published
|
|
6UB9
 
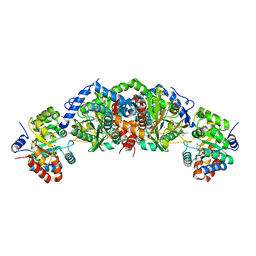 | | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM | | 分子名称: | (2R,3S,4R)-3-(4'-chloro-2',6'-difluoro[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | 著者 | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-09-11 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.783 Å) | | 主引用文献 | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM
To be Published
|
|
6UAH
 
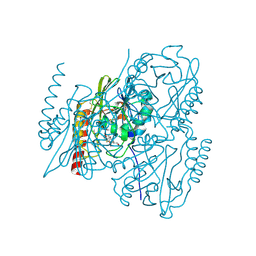 | | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem | | 分子名称: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-09-10 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem
To Be Published
|
|
6V3Q
 
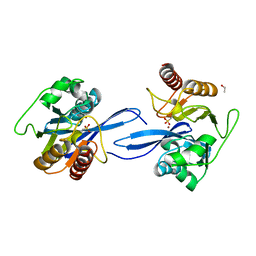 | | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form | | 分子名称: | ISOPROPYL ALCOHOL, Metallo-beta-lactamase FIM-1, ZINC ION | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-11-26 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form
To Be Published
|
|
6V5M
 
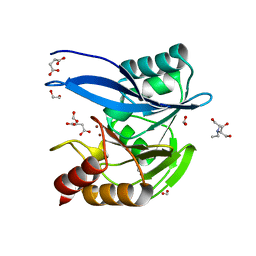 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | 著者 | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-04 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate.
To Be Published
|
|
6V71
 
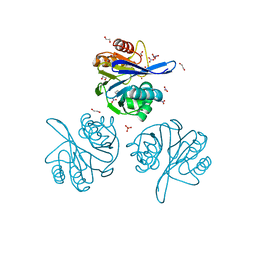 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID, ... | | 著者 | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-06 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site
To Be Published
|
|
6W33
 
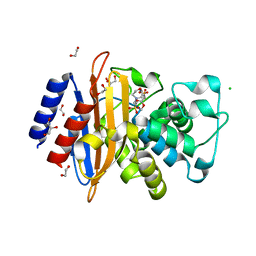 | | Crystal Structure of Class A Beta-lactamase from Bacillus cereus in the Complex with the Beta-lactamase Inhibitor Clavulanate | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-08 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of Class A Beta-lactamase from
Bacillus cereus in the Complex with the Beta-lactamase Inhibitor Clavulanate
To Be Published
|
|
7K1O
 
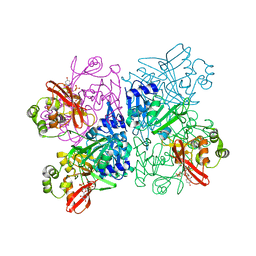 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate | | 分子名称: | 1,2-ETHANEDIOL, 1-(3,5-di-O-phosphono-alpha-L-xylofuranosyl)pyrimidine-2,4(1H,3H)-dione, Uridylate-specific endoribonuclease | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Welk, L, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-08 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate
To Be Published
|
|
7KOJ
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide, ACETATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-09 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494
to be published
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-09 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
7KOK
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-09 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496
to be published
|
|
3UUW
 
 | | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile. | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-11-28 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile.
TO BE PUBLISHED
|
|
7KP2
 
 | | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin | | 分子名称: | L-NEOPTERIN, Putative Pterin Binding Protein | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Pshenychnyi, S, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-10 | | 公開日 | 2021-11-17 | | 最終更新日 | 2022-08-17 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin.
To Be Published
|
|
3VCZ
 
 | | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6 | | 分子名称: | CALCIUM ION, Endoribonuclease L-PSP, GLYCEROL, ... | | 著者 | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-01-04 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6
To be Published
|
|
6DIN
 
 | | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase | | 分子名称: | SULFATE ION, dTDP-4-dehydrorhamnose 3,5-epimerase | | 著者 | Chang, C, Jedrzejczak, R, Chhor, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-05-23 | | 公開日 | 2018-05-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase
To Be Published
|
|
