1SLA
 
 | |
1SLC
 
 | | X-RAY CRYSTALLOGRAPHY REVEALS CROSSLINKING OF MAMMALIAN LECTIN (GALECTIN-1) BY BIANTENNARY COMPLEX TYPE SACCHARIDES | | Descriptor: | BOVINE GALECTIN-1, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bourne, Y, Cambillau, C. | | Deposit date: | 1994-03-12 | | Release date: | 1995-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides.
Nat.Struct.Biol., 1, 1994
|
|
1HZU
 
 | | DOMAIN SWING UPON HIS TO ALA MUTATION IN NITRITE REDUCTASE OF PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRITE REDUCTASE | | Authors: | Brown, K, Cutruzzola, F, Brunori, M, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-01-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Domain swing upon His to Ala mutation in nitrite reductase of Pseudomonas aeruginosa.
J.Mol.Biol., 312, 2001
|
|
1HZV
 
 | | DOMAIN SWING UPON HIS TO ALA MUTATION IN NITRITE REDUCTASE OF PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-01-26 | | Release date: | 2001-09-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Domain swing upon His to Ala mutation in nitrite reductase of Pseudomonas aeruginosa.
J.Mol.Biol., 312, 2001
|
|
1I3U
 
 | | THREE-DIMENSIONAL STRUCTURE OF A LLAMA VHH DOMAIN COMPLEXED WITH THE DYE RR1 | | Descriptor: | 5-(4,6-DIAMINO-[1,3,5]TRIAZIN-2-YLAMINO)-4-HYDROXY-3-(2-SULFO-PHENYLAZO)-NAPHTHALENE-2,7-DISULFONIC ACID, ANTIBODY VHH LAMA DOMAIN, SULFATE ION | | Authors: | Spinelli, S, Tegoni, M, Frenken, L, van Vliet, C, Cambillau, C. | | Deposit date: | 2001-02-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lateral recognition of a dye hapten by a llama VHH domain.
J.Mol.Biol., 311, 2001
|
|
1I3V
 
 | | THREE-DIMENSIONAL STRUCTURE OF A LAMA VHH DOMAIN UNLIGANDED | | Descriptor: | ANTIBODY VHH LAMA DOMAIN | | Authors: | Spinelli, S, Tegoni, M, Frenken, L, van Vliet, C, Cambillau, C. | | Deposit date: | 2001-02-16 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lateral recognition of a dye hapten by a llama VHH domain.
J.Mol.Biol., 311, 2001
|
|
1K19
 
 | | NMR Solution Structure of the Chemosensory Protein CSP2 from Moth Mamestra brassicae | | Descriptor: | Chemosensory Protein CSP2 | | Authors: | Mosbah, A, Campanacci, V, Lartigue, A, Tegoni, M, Cambillau, C, Darbon, H. | | Deposit date: | 2001-09-24 | | Release date: | 2002-12-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a chemosensory protein from the moth Mamestra brassicae
BIOCHEM.J., 369, 2003
|
|
1G1K
 
 | | COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDING PROTEIN | | Authors: | Spinelli, S, Fierobe, H.-P, Belaich, A, Belaich, J.-P, Henrissat, B, Cambillau, C. | | Deposit date: | 2000-10-12 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cohesin module from Clostridium cellulolyticum: implications for dockerin recognition.
J.Mol.Biol., 304, 2000
|
|
1LGB
 
 | |
1LOB
 
 | |
1LOA
 
 | |
1SJV
 
 | | Three-Dimensional Structure of a Llama VHH Domain Swapping | | Descriptor: | r9 | | Authors: | Spinelli, S, Desmyter, A, Frenken, L, Verrips, T, Cambillau, C. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Domain swapping of a llama VHH domain builds a crystal-wide beta-sheet structure.
Febs Lett., 564, 2004
|
|
1U08
 
 | | Crystal Structure and Reactivity of YbdL from Escherichia coli Identify a Methionine Aminotransferase Function. | | Descriptor: | Hypothetical aminotransferase ybdL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dolzan, M, Johansson, K, Roig-Zamboni, V, Campanacci, V, Tegoni, M, Schneider, G, Cambillau, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function
FEBS Lett., 571, 2004
|
|
1W9A
 
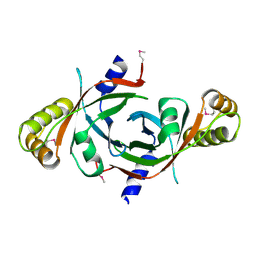 | | Crystal structure of Rv1155 from Mycobacterium tuberculosis | | Descriptor: | PUTATIVE PYRIDOXINE/PYRIDOXAMINE 5'-PHOSPHATE OXIDASE | | Authors: | Cannan, S, Sulzenbacher, G, Roig-Zamboni, V, Scappuccini, L, Frassinetti, F, Maurien, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2004-10-07 | | Release date: | 2005-01-06 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Rv1155 from Mycobacterium Tuberculosis
FEBS Lett., 579, 2005
|
|
1HN2
 
 | | CRYSTAL STRUCTURE OF BOVINE OBP COMPLEXED WITH AMINOANTHRACENE | | Descriptor: | (3R)-oct-1-en-3-ol, ANTHRACEN-1-YLAMINE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
1LOE
 
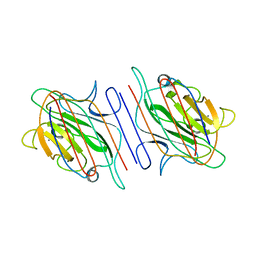 | |
3BFH
 
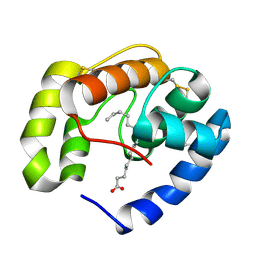 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with hexadecanoic acid | | Descriptor: | CHLORIDE ION, PALMITIC ACID, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3BFB
 
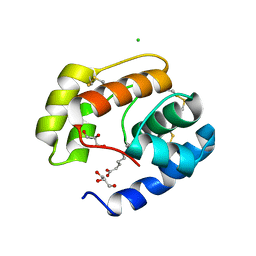 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the 9-keto-2(E)-decenoic acid | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3BFA
 
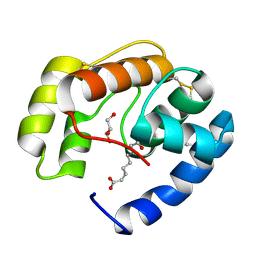 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the Queen mandibular pheromone | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3CAB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 7.0 | | Descriptor: | GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-19 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3CDN
 
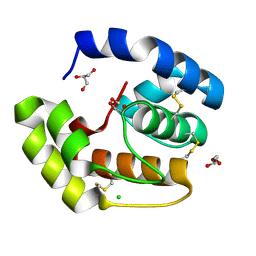 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 4.0 | | Descriptor: | CHLORIDE ION, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3CZ1
 
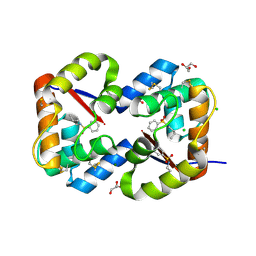 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with the n-butyl benzene sulfonamide at pH 7.0 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3CYZ
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with 9-keto-2(E)-decenoic acid at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3CZ0
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with the n-butyl benzene sulfonamide at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3CZ2
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera at pH 7.0 | | Descriptor: | CHLORIDE ION, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
