7NFI
 
 | |
7NFJ
 
 | |
7NFO
 
 | |
7NFK
 
 | | An octameric barrel state of a de novo coiled-coil assembly: CC-Type2-(LaId)4-I24S. | | Descriptor: | CC-Type2-(LaId)4-I24S, ISOPROPYL ALCOHOL | | Authors: | Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
7NFP
 
 | | A heptameric barrel state of a de novo coiled-coil assembly: CC-Type2-(LaId)4-I17K | | Descriptor: | CC-Type2-(LaId)4-I17K, GLYCEROL | | Authors: | Burton, A.J, Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
6ZT1
 
 | |
7A1T
 
 | | A collapsed hexameric state of a de novo coiled-coil assembly: CC-Type2-(GgLaId)4-W19BrPhe. | | Descriptor: | ACETATE ION, CADMIUM ION, CC-Type2-(GgLaId)4-W19BrPhe, ... | | Authors: | Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-05-19 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Coiled coils 9-to-5: rational de novo design of alpha-helical barrels with tunable oligomeric states.
Chem Sci, 12, 2021
|
|
2PET
 
 | | Lutheran glycoprotein, N-terminal domains 1 and 2. | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-03 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
2PF6
 
 | | Lutheran glycoprotein, N-terminal domains 1 and 2 | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-04 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
2PLX
 
 | |
2QYQ
 
 | |
2QIH
 
 | |
6Q5S
 
 | | Crystal structure of a de novo designed antiparallel four-helix coiled coil apCC-Tet | | Descriptor: | apCC-Tet | | Authors: | Beesley, J.L, Guto, G.R, Wood, C.W, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5I
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24E | | Descriptor: | AMMONIUM ION, CC-Hex*-L24E, SULFATE ION | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5L
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24H | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CC-Hex*-L24H | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5P
 
 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-II | | Descriptor: | CC-Hex*-II, GLYCEROL, PHOSPHATE ION | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5O
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-LL | | Descriptor: | CC-Hex*-LL | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
2X8L
 
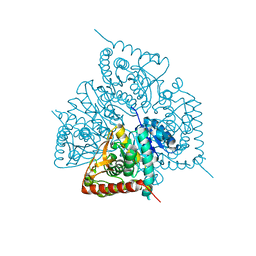 | |
2WWR
 
 | | Crystal Structure of Human Glyoxylate Reductase Hydroxypyruvate Reductase | | Descriptor: | GLYOXYLATE REDUCTASE/HYDROXYPYRUVATE REDUCTASE, MAGNESIUM ION | | Authors: | Booth, M.P.S, Conners, R, Rumsby, G, Brady, R.L. | | Deposit date: | 2009-10-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis of Substrate Specificity in Human Glyoxylate Reductase/Hydroxypyruvate Reductase.
J.Mol.Biol., 360, 2006
|
|
1OC4
 
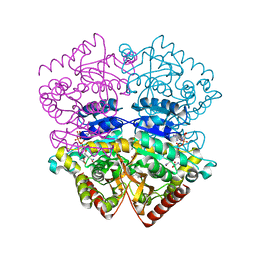 | | Lactate dehydrogenase from Plasmodium berghei | | Descriptor: | GLYCEROL, L-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Winter, V.J, Brady, R.L. | | Deposit date: | 2003-02-05 | | Release date: | 2003-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Plasmodium Berghei Lactate Dehydrogenase Indicates the Unique Structural Differences of These Enzymes are Shared Across the Plasmodium Genus
Mol.Biochem.Parasitol., 131, 2003
|
|
1SFI
 
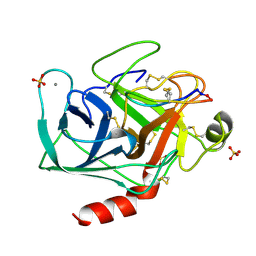 | | High resolution structure of a potent, cyclic protease inhibitor from sunflower seeds | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN, ... | | Authors: | Luckett, S, Garcia, R.S, Barker, J.J, Konarev, A.V, Shewry, P, Clarke, A.R, Brady, R.L. | | Deposit date: | 1998-12-16 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds.
J.Mol.Biol., 290, 1999
|
|
1I0Z
 
 | | HUMAN HEART L-LACTATE DEHYDROGENASE H CHAIN, TERNARY COMPLEX WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE H CHAIN, OXAMIC ACID | | Authors: | Read, J.A, Winter, V.J, Eszes, C.M, Sessions, R.B, Brady, R.L. | | Deposit date: | 2001-01-30 | | Release date: | 2001-03-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase.
Proteins, 43, 2001
|
|
1I10
 
 | | HUMAN MUSCLE L-LACTATE DEHYDROGENASE M CHAIN, TERNARY COMPLEX WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-LACTATE DEHYDROGENASE M CHAIN, ... | | Authors: | Read, J.A, Winter, V.J, Eszes, C.M, Sessions, R.B, Brady, R.L. | | Deposit date: | 2001-01-30 | | Release date: | 2001-03-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase.
Proteins, 43, 2001
|
|
1KN3
 
 | |
1CET
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
