4NRU
 
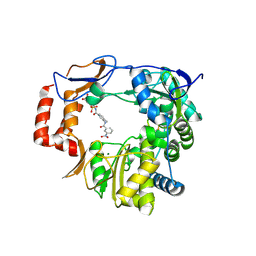 | | Murine Norovirus RNA-dependent-RNA-polymerase in complex with Compound 6, a suramin derivative | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, MAGNESIUM ION, RNA dependent RNA polymerase | | Authors: | Milani, M, Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
1RTE
 
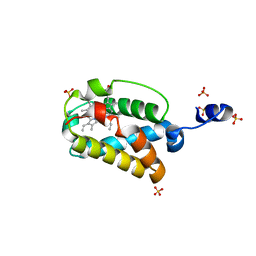 | | X-ray Structure of Cyanide Derivative of Truncated Hemoglobin N (trHbN) from Mycobacterium Tuberculosis | | Descriptor: | CYANIDE ION, Hemoglobin-like protein HbN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Milani, M, Guertin, M, Boffi, A, Antonini, G, Bocedi, A, Mattu, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 2003-12-10 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyanide binding to truncated hemoglobins: a crystallographic and kinetic study
Biochemistry, 43, 2004
|
|
2QEQ
 
 | |
1T1V
 
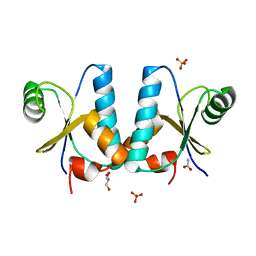 | | Crystal Structure of the Glutaredoxin-like Protein SH3BGRL3 at 1.6 A resolution | | Descriptor: | ACETIC ACID, GLYCEROL, SH3 domain-binding glutamic acid-rich protein-like 3, ... | | Authors: | Nardini, M, Mazzocco, M, Massaro, M, Maffei, M, Vergano, A, Donadini, A, Scartezzini, M, Bolognesi, M. | | Deposit date: | 2004-04-19 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the glutaredoxin-like protein SH3BGRL3 at 1.6 A resolution
Biochem.Biophys.Res.Commun., 318, 2004
|
|
1HL3
 
 | | CtBP/BARS in ternary complex with NAD(H) and PIDLSKK peptide | | Descriptor: | C-TERMINAL BINDING PROTEIN 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PRO-ILE-ASP-LEU-SER-LYS-LYS PEPTIDE | | Authors: | Nardini, M, Spano, S, Cericola, C, Pesce, A, Massaro, A, Millo, E, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2003-03-13 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ctbp/Bars: A Dual-Function Protein Involved in Transcription Co-Repression and Golgi Membrane Fission
Embo J., 22, 2003
|
|
2VYY
 
 | | Mutant Ala55Trp of Cerebratuls lacteus mini-hemoglobin | | Descriptor: | GLYCEROL, NEURAL HEMOGLOBIN, OXYGEN MOLECULE, ... | | Authors: | Salter, M.D, Nienhaus, K, Nienhaus, G.U, Dewilde, S, Moens, L, Pesce, A, Nardini, M, Bolognesi, M, Olson, J.S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Apolar Channel in Cerebratulus Lacteus Hemoglobin is the Route for O2 Entry and Exit.
J.Biol.Chem., 283, 2008
|
|
2VYZ
 
 | | Mutant Ala55Phe of Cerebratulus lacteus mini-hemoglobin | | Descriptor: | ACETATE ION, GLYCEROL, NEURAL HEMOGLOBIN, ... | | Authors: | Salter, M.D, Nienhaus, K, Nienhaus, G.U, Dewilde, S, Moens, L, Pesce, A, Nardini, M, Bolognesi, M, Olson, J.S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Apolar Channel in Cerebratulus Lacteus Hemoglobin is the Route for O2 Entry and Exit.
J.Biol.Chem., 283, 2008
|
|
2VYW
 
 | | Hemoglobin (Hb2) from trematode Fasciola hepatica | | Descriptor: | HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dewilde, S, Ioanitescu, A.I, Kiger, L, Gilany, K, Marden, M.C, Van Doorslaer, S, Vercruysse, J, Pesce, A, Nardini, M, Bolognesi, M, Moens, L. | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hemoglobins of the Trematodes Fasciola Hepatica and Paramphistomum Epiclitum: A Molecular Biological, Physico-Chemical, Kinetic, and Vaccination Study.
Protein Sci., 17, 2008
|
|
7ZPA
 
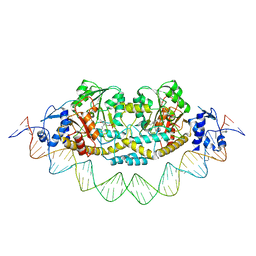 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
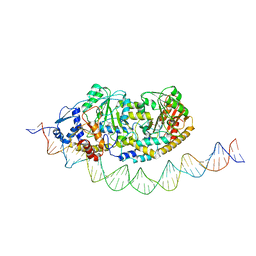 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
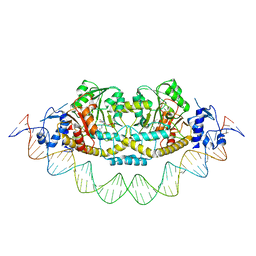 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZTH
 
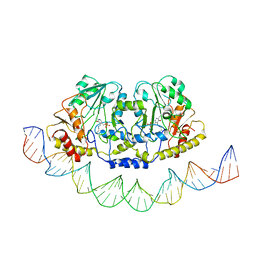 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
1URR
 
 | |
2XKG
 
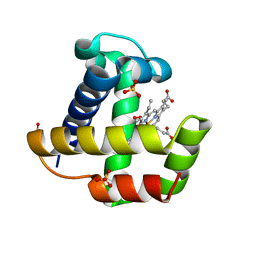 | | C.lacteus mini-Hb Leu86Ala mutant | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2XKI
 
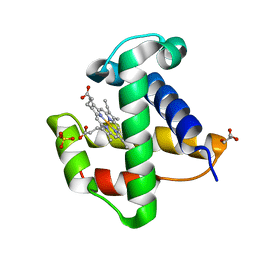 | | Aquo-met structure of C.lacteus mini-Hb | | Descriptor: | GLYCEROL, NEURAL HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2XKH
 
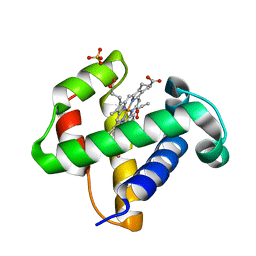 | | Xe derivative of C.lacteus mini-Hb Leu86Ala mutant | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2XHZ
 
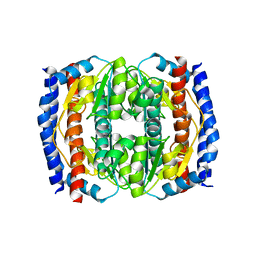 | | Probing the active site of the sugar isomerase domain from E. coli arabinose-5-phosphate isomerase via X-ray crystallography | | Descriptor: | ARABINOSE 5-PHOSPHATE ISOMERASE | | Authors: | Gourlay, L.J, Sommaruga, S, Nardini, M, Sperandeo, P, Deho, G, Polissi, A, Bolognesi, M. | | Deposit date: | 2010-06-24 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing the Active Site of the Sugar Isomerase Domain from E. Coli Arabinose-5-Phosphate Isomerase Via X-Ray Crystallography.
Protein Sci., 19, 2010
|
|
1KR7
 
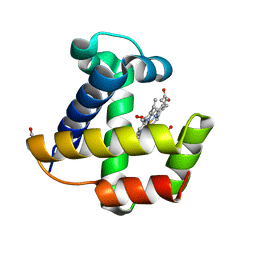 | | Crystal structure of the nerve tissue mini-hemoglobin from the nemertean worm Cerebratulus lacteus | | Descriptor: | ACETATE ION, Neural globin, OXYGEN MOLECULE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Geuens, E, Yamauchi, k, Ascenzi, P, Riggs, A.F, Moens, L, Bolognesi, M. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 109 residue nerve tissue minihemoglobin from Cerebratulus lacteus highlights striking structural plasticity of the alpha-helical globin fold
Structure, 10, 2002
|
|
2VG7
 
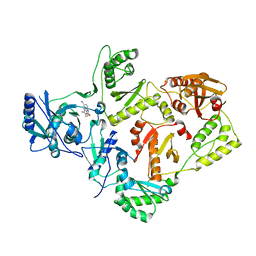 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-iodophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
2VG6
 
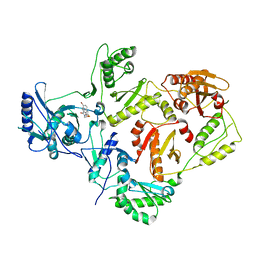 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-bromophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
2VG5
 
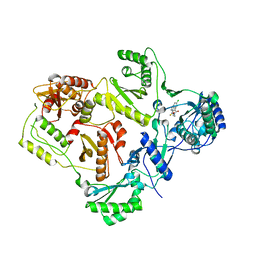 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-chlorophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
6S6U
 
 | |
4AVD
 
 | | C.lacteus nerve Hb in complex with CO | | Descriptor: | ACETATE ION, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Germani, F, Pesce, A, Venturini, A, Moens, L, Bolognesi, M, Dewilde, S, Nardini, M. | | Deposit date: | 2012-05-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Crystal Structures of the Cerebratulus Lacteus Mini-Hb in the Unligated and Carbomonoxy States.
Int.J.Mol.Sci., 13, 2012
|
|
4B5C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia pseudomallei | | Descriptor: | ACETATE ION, PUTATIVE OMPA FAMILY LIPOPROTEIN | | Authors: | Gourlay, L.J, Peri, C, Conchillo-Sole, O, Ferrer-Navarro, M, Gori, A, Longhi, R, Rinchai, D, Lertmemongkolchai, G, Lassaux, P, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Burkholderia Pseudomallei Acute Phase Antigen Bpsl2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology.
Chem.Biool., 20, 2013
|
|
6S6S
 
 | |
