1MFZ
 
 | |
1MV8
 
 | | 1.55 A crystal structure of a ternary complex of GDP-mannose dehydrogenase from Psuedomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, GDP-mannose 6-dehydrogenase, ... | | Authors: | Snook, C.F, Tipton, P.A, Beamer, L.J. | | Deposit date: | 2002-09-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of GDP-mannose
dehydrogenase: A key enzyme in alginate
biosynthesis of P. aeruginosa
Biochemistry, 42, 2003
|
|
1MUU
 
 | | 2.0 A crystal structure of GDP-mannose dehydrogenase | | Descriptor: | GDP-mannose 6-dehydrogenase, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), P'-D-MANNOPYRANOSYL ESTER, ... | | Authors: | Snook, C.F, Tipton, P.A, Beamer, L.J. | | Deposit date: | 2002-09-24 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of GDP-mannose dehydrogenase: A key enzyme of alginate biosynthesis in P. aeruginosa
Biochemistry, 42, 2003
|
|
7S77
 
 | | Crystal structure of the G391V variant of human PGM-1 | | Descriptor: | Phosphoglucomutase-1, SULFATE ION | | Authors: | Stiers, K.M, Beamer, L.J. | | Deposit date: | 2021-09-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Effects of the T337M and G391V disease-related variants on human phosphoglucomutase 1: structural disruptions large and small.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
1P5D
 
 | |
1P5G
 
 | |
1PCJ
 
 | |
1PCM
 
 | |
3PDK
 
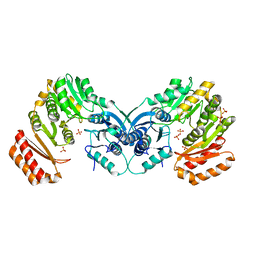 | | crystal structure of phosphoglucosamine mutase from B. anthracis | | Descriptor: | PHOSPHATE ION, Phosphoglucosamine mutase | | Authors: | Mehra-Chaudhary, R, Mick, J, Tanner, J.J, Henzl, M, Beamer, L.J. | | Deposit date: | 2010-10-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Bacillus anthracis Phosphoglucosamine Mutase, an Enzyme in the Peptidoglycan Biosynthetic Pathway.
J.Bacteriol., 193, 2011
|
|
6UIQ
 
 | |
6UXK
 
 | |
6UXI
 
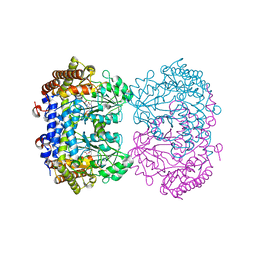 | | Structure of serine hydroxymethyltransferase 8 from Glycine max cultivar Essex complexed with PLP-Glycine | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase | | Authors: | Korasick, D.A, Tanner, J.J, Beamer, L.J. | | Deposit date: | 2019-11-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Impaired folate binding of serine hydroxymethyltransferase 8 from soybean underlies resistance to the soybean cyst nematode.
J.Biol.Chem., 295, 2020
|
|
6UXJ
 
 | | Structure of serine hydroxymethyltransferase 8 from Glycine max cultivar Essex complexed with PLP-glycine and 5-formyltetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Korasick, D.A, Tanner, J.J, Beamer, L.J. | | Deposit date: | 2019-11-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impaired folate binding of serine hydroxymethyltransferase 8 from soybean underlies resistance to the soybean cyst nematode.
J.Biol.Chem., 295, 2020
|
|
6UXH
 
 | |
6UXL
 
 | |
1K2Y
 
 | |
1K35
 
 | |
5VBI
 
 | |
5VG7
 
 | |
5VIN
 
 | |
5VEC
 
 | |
6NOL
 
 | |
6NN2
 
 | | Xanthomonas citri PGM Apo-Phospho | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Phosphoglucomutase | | Authors: | Stiers, K.M, Beamer, L.J. | | Deposit date: | 2019-01-14 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural and dynamical description of the enzymatic reaction of a phosphohexomutase.
Struct Dyn., 6, 2019
|
|
6NNT
 
 | |
6NN1
 
 | | Xanthomonas citri PGM Apo-Dephospho | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stiers, K.M, Beamer, L.J. | | Deposit date: | 2019-01-14 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and dynamical description of the enzymatic reaction of a phosphohexomutase.
Struct Dyn., 6, 2019
|
|
