5LZP
 
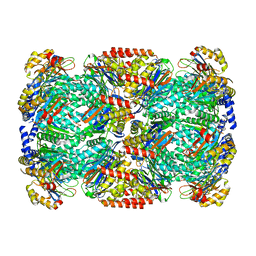 | | Binding of the C-terminal GQYL motif of the bacterial proteasome activator Bpa to the 20S proteasome | | Descriptor: | Bacterial proteasome activator, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
5LRT
 
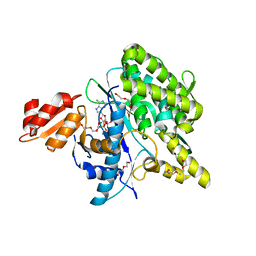 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP and Phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Depupylase, ... | | Authors: | Bolten, M, Vahlensieck, C, Lipp, C, Leibundgut, M, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-08-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Depupylase Dop Requires Inorganic Phosphate in the Active Site for Catalysis.
J. Biol. Chem., 292, 2017
|
|
4V59
 
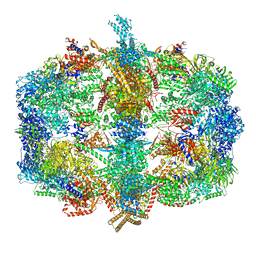 | | Crystal structure of fatty acid synthase complexed with nadp+ from thermomyces lanuginosus at 3.1 angstrom resolution. | | Descriptor: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | Deposit date: | 2007-03-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
4V5O
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH INITIATION FACTOR 1. | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S12, 40S RIBOSOMAL PROTEIN S3A, ... | | Authors: | Rabl, J, Leibundgut, M, Ataide, S.F, Haag, A, Ban, N. | | Deposit date: | 2010-11-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1.
Science, 331, 2011
|
|
4V8P
 
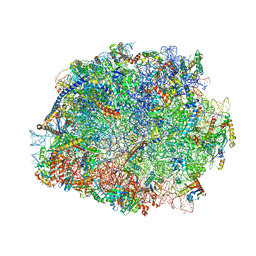 | | T.thermophila 60S ribosomal subunit in complex with initiation factor 6. | | Descriptor: | 26S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Klinge, S, Voigts-Hoffmann, F, Leibundgut, M, Arpagaus, S, Ban, N. | | Deposit date: | 2011-09-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6.
Science, 334, 2011
|
|
4V58
 
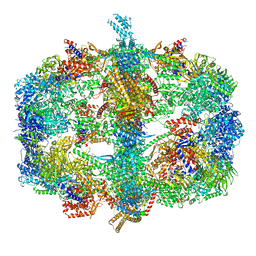 | | Crystal structure of fatty acid synthase from thermomyces lanuginosus at 3.1 angstrom resolution. | | Descriptor: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE | | Authors: | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | Deposit date: | 2007-03-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
4V8L
 
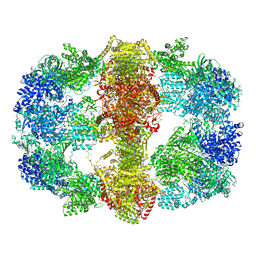 | |
4V8T
 
 | | Cryo-EM Structure of the 60S Ribosomal Subunit in Complex with Arx1 and Rei1 | | Descriptor: | 25S RIBOSOMAL RNA, 5.8S RIBOSOMAL RNA, 5S RIBOSOMAL RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Montellese, C, Ban, N. | | Deposit date: | 2012-08-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Cryo-Em Structures of Arx1 and Maturation Factors Rei1 and Jjj1 Bound to the 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 19, 2012
|
|
4V5B
 
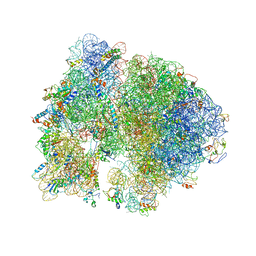 | | Structure of PDF binding helix in complex with the ribosome. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Bingel-Erlenmeyer, R, Kohler, R, Kramer, G, Sandikci, A, Antolic, S, Maier, T, Schaffitzel, C, Wiedmann, B, Bukau, B, Ban, N. | | Deposit date: | 2007-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | A Peptide Deformylase-Ribosome Complex Reveals Mechanism of Nascent Chain Processing.
Nature, 452, 2008
|
|
2VZ8
 
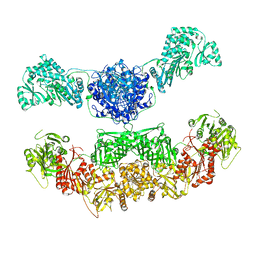 | |
2VZ9
 
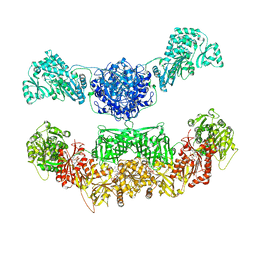 | |
1K9M
 
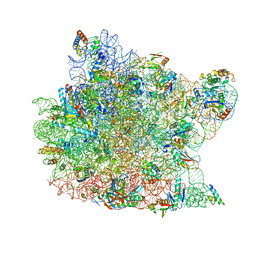 | | Co-crystal structure of tylosin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-10-29 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1KC8
 
 | | Co-crystal Structure of Blasticidin S Bound to the 50S Ribosomal Subunit | | Descriptor: | 23S RRNA, 5S RRNA, BLASTICIDIN S, ... | | Authors: | Hansen, J.L, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-11-07 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
1KD1
 
 | | Co-crystal Structure of Spiramycin bound to the 50S Ribosomal Subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-11-12 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1M1K
 
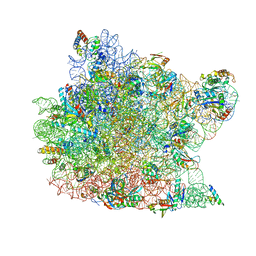 | | Co-crystal structure of azithromycin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, AZITHROMYCIN, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-06-19 | | Release date: | 2002-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
6FAI
 
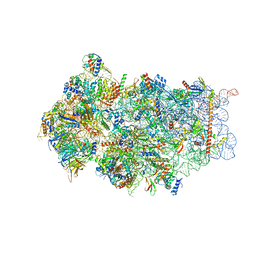 | | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Scaiola, A, Pena, C, Weisser, M, Boehringer, D, Leibundgut, M, Klingauf-Nerurkar, P, Gerhardy, S, Panse, V.G, Ban, N. | | Deposit date: | 2017-12-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit.
EMBO J., 37, 2018
|
|
4B0S
 
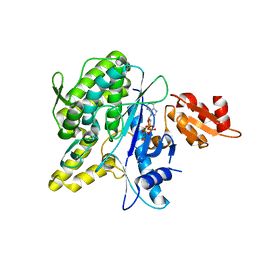 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEAMIDASE-DEPUPYLASE DOP, MAGNESIUM ION | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
6FRK
 
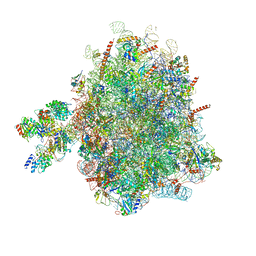 | | Structure of a prehandover mammalian ribosomal SRP and SRP receptor targeting complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kobayashi, K, Jomaa, A, Ban, N. | | Deposit date: | 2018-02-16 | | Release date: | 2018-03-28 | | Last modified: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a prehandover mammalian ribosomal SRP·SRP receptor targeting complex.
Science, 360, 2018
|
|
4B0R
 
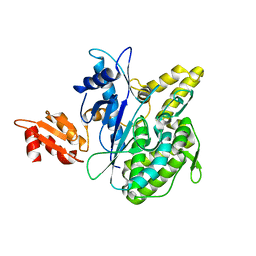 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway | | Descriptor: | DEAMIDASE-DEPUPYLASE DOP | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Pup ligase PafA and depupylase Dop from the prokaryotic ubiquitin-like modification pathway.
Nat Commun, 3, 2012
|
|
4B0T
 
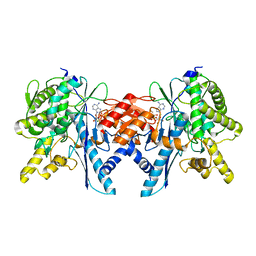 | | Structure of the Pup Ligase PafA of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
6GB2
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 39S ribosomal subunit. | | Descriptor: | 'Mitochondrial ribosomal protein L30, 'Mitochondrial ribosomal protein L55, 'Mitochondrial ribosomal protein L59, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
8OIP
 
 | | 28S mammalian mitochondrial small ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 28S ribosomal protein S15, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
8OIS
 
 | | 28S human mitochondrial small ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
8OIR
 
 | | 55S human mitochondrial ribosome with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
8OIT
 
 | | 39S human mitochondrial large ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 16S rRNA, 39S ribosomal protein L1, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
