4ZVH
 
 | |
2GGE
 
 | | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, yitF | | Authors: | Malashkevich, V.N, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A
To be Published
|
|
5I6E
 
 | | Crystal structure of the central domain of yeast acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase, MALONATE ION | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
3MAE
 
 | | CRYSTAL STRUCTURE OF PROBABLE DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM LISTERIA MONOCYTOGENES 4b F2365 | | Descriptor: | 2-oxoisovalerate dehydrogenase E2 component, dihydrolipoamide acetyltransferase, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A CATALYTIC DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM LISTERIA MONOCYTOGENES 4b F2365
To be Published
|
|
3VC2
 
 | | Crystal structure of geranyl diphosphate C-methyltransferase from Streptomyces coelicolor A3(2) in complex with Mg2+, geranyl diphosphate, and S-adenosyl-L-homocysteine | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2012-01-03 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Structure of Geranyl Diphosphate C-Methyltransferase from Streptomyces coelicolor and Implications for the Mechanism of Isoprenoid Modification.
Biochemistry, 51, 2012
|
|
3VM0
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - NO2 complex from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
5I36
 
 | | Crystal structure of color device state A | | Descriptor: | DNA (26-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*GP*TP*GP*GP*TP*AP*TP*C)-3'), DNA (5'-D(*CP*AP*GP*AP*TP*AP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Hao, Y, Kristiansen, M, Sha, R, Birktoft, J, Mao, C, Seeman, N.C. | | Deposit date: | 2016-02-09 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.123 Å) | | Cite: | A device that operates within a self-assembled 3D DNA crystal.
Nat Chem, 9, 2017
|
|
3MBV
 
 | | Structure of bacterirhodopsin crystallized in betta-XylOC(16+4) meso phase | | Descriptor: | (3R,7R,11R)-3,7,11,15-tetramethylhexadecyl alpha-D-ribopyranoside, Bacteriorhodopsin, RETINAL | | Authors: | Borshchevskiy, V, Moiseeva, E, Kuklin, A, Bueldt, G, Hato, M, Gordeliy, V. | | Deposit date: | 2010-03-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoprenoid-chained lipid beta-XylOC16+4 A novel molecule for in meso membrane protein crystallization
J.Cryst.Growth, 312, 2010
|
|
2GEF
 
 | |
4ZU7
 
 | | X-ray structure if the QdtA 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum, double mutant Y17R/R97H, in complex with TDP | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, QdtA, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
5A0R
 
 | | Product peptide-bound structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | Descriptor: | GLYCEROL, PRODUCT PEPTIDE, ZINC ION, ... | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
3MD3
 
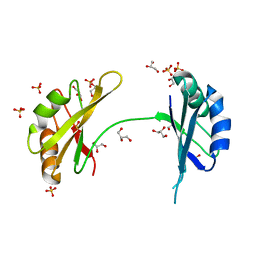 | | Crystal Structure of the First Two RRM Domains of Yeast Poly(U) Binding Protein (Pub1) | | Descriptor: | GLYCEROL, Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1, SULFATE ION | | Authors: | Li, H, Shi, H, Zhu, Z, Wang, H, Niu, L, Teng, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the First Two RRM Domains of Yeast Poly(U) Binding Protein (Pub1)
To be published
|
|
3MDD
 
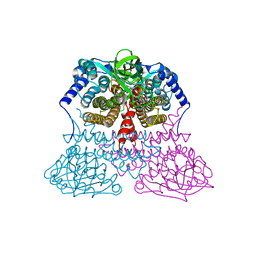 | |
3VEJ
 
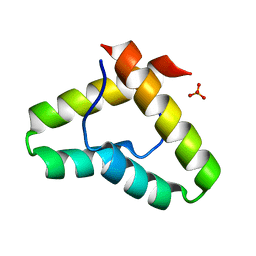 | | Crystal structure of the Get5 carboxyl domain from S. cerevisiae | | Descriptor: | PHOSPHATE ION, Ubiquitin-like protein MDY2 | | Authors: | Chartron, J.W, Vandervelde, D.G, Rao, M, Clemons Jr, W.M. | | Deposit date: | 2012-01-08 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Get5 Carboxyl-terminal Domain Is a Novel Dimerization Motif That Tethers an Extended Get4/Get5 Complex.
J.Biol.Chem., 287, 2012
|
|
5A1W
 
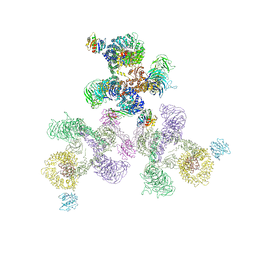 | | The structure of the COPI coat linkage II | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A3S
 
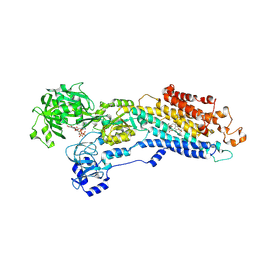 | | Crystal structure of the (SR) Calcium ATPase E2-vanadate complex bound to thapsigargin and TNP-ATP | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Clausen, J.D, Bublitz, M, Arnou, B, Olesen, C, Andersen, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2015-06-03 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Structure, 24, 2016
|
|
4ZVE
 
 | | Crystal structure of GGDEF domain of the E. coli DosC - form I (apo-form) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Diguanylate cyclase DosC | | Authors: | Tarnawski, M, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2015-05-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analysis of an oxygen-regulated diguanylate cyclase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VR0
 
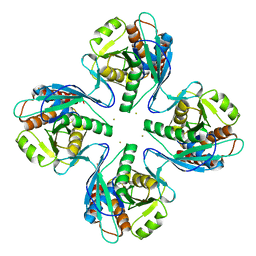 | | Crystal structure of Pyrococcus furiosus PbaB, an archaeal proteasome activator | | Descriptor: | GOLD ION, Putative uncharacterized protein | | Authors: | Kumoi, K, Satoh, T, Hiromoto, T, Mizushima, T, Kamiya, Y, Noda, M, Uchiyama, S, Murata, K, Yagi, H, Kato, K. | | Deposit date: | 2012-04-02 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An archaeal homolog of proteasome assembly factor functions as a proteasome activator
Plos One, 8, 2013
|
|
3MFQ
 
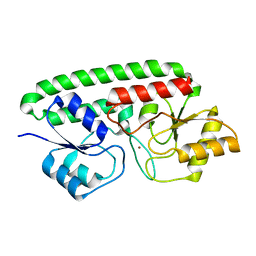 | | A Glance into the Metal Binding Specificity of TroA: Where Elaborate Behaviors Occur in the Active Center | | Descriptor: | High-affinity zinc uptake system protein znuA, ZINC ION | | Authors: | Gao, G.F, Zheng, B, Zhang, Q, Gao, J, Han, H, Li, M. | | Deposit date: | 2010-04-03 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Insight into the interaction of metal ions with TroA from Streptococcus suis
Plos One, 6, 2011
|
|
2G8F
 
 | |
5A95
 
 | | Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
2G8U
 
 | |
3VG6
 
 | | Barium derivative of human LFABP | | Descriptor: | BARIUM ION, Fatty acid-binding protein, liver, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
2GBL
 
 | | Crystal Structure of Full Length Circadian Clock Protein KaiC with Phosphorylation Sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.R, Pattanayek, S, Xu, Y, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2006-03-10 | | Release date: | 2007-01-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods.
Embo J., 25, 2006
|
|
3VGJ
 
 | | Crystal of Plasmodium falciparum tyrosyl-tRNA synthetase (PfTyrRS)in complex with adenylate analog | | Descriptor: | ADENOSINE MONOPHOSPHATE, TYROSINE, Tyrosyl-tRNA synthetase, ... | | Authors: | Banday, M.M, Yogavel, M, Bhatt, T.K, Khan, S, Sharma, A, Sharma, A. | | Deposit date: | 2011-08-14 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Malaria parasite tyrosyl-tRNA synthetase secretion triggers pro-inflammatory responses.
Nat Commun, 2, 2011
|
|
