3NJF
 
 | | Y26F mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJN
 
 | | Q118A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJM
 
 | | P117A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJG
 
 | | K98A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJK
 
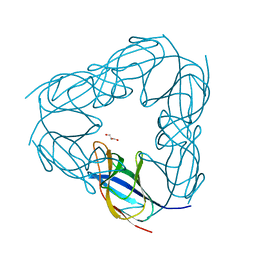 | | D116A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis, at pH5.5 | | Descriptor: | GLYCEROL, Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3DN7
 
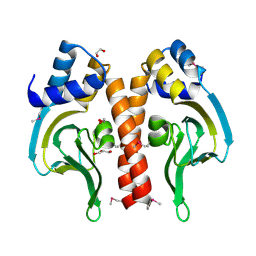 | | Cyclic nucleotide binding regulatory protein from Cytophaga hutchinsonii. | | Descriptor: | 1,2-ETHANEDIOL, Cyclic nucleotide binding regulatory protein, GLYCEROL | | Authors: | Osipiuk, J, Maltseva, N, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of cyclic nucleotide binding regulatory protein from Cytophaga hutchinsonii.
To be Published
|
|
3O6D
 
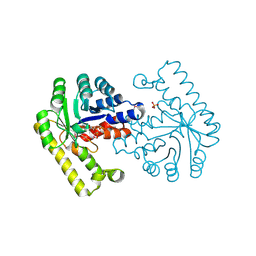 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni in complex with pyridoxine-5'-phosphate | | Descriptor: | PHOSPHATE ION, PYRIDOXINE-5'-PHOSPHATE, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
3FDX
 
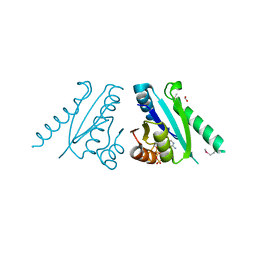 | | Putative filament protein / universal stress protein F from Klebsiella pneumoniae. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | X-ray crystal structure of putative filament protein / universal stress protein F from Klebsiella pneumoniae.
To be Published
|
|
3O6C
 
 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
3G0S
 
 | | Dihydrodipicolinate synthase from Salmonella typhimurium LT2 | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Osipiuk, J, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structure of dihydrodipicolinate synthase from Salmonella typhimurium LT2.
To be Published
|
|
3GBX
 
 | | Serine hydroxymethyltransferase from Salmonella typhimurium | | Descriptor: | ACETATE ION, Serine hydroxymethyltransferase | | Authors: | Osipiuk, J, Nocek, B, Zhou, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of serine hydroxymethyltransferase from Salmonella typhimurium.
To be Published
|
|
3OMS
 
 | | Putative 3-demethylubiquinone-9 3-methyltransferase, PhnB protein, from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, PhnB protein | | Authors: | Osipiuk, J, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of putative 3-demethylubiquinone-9
3-methyltransferase, PhnB protein, from Bacillus cereus.
To be Published
|
|
3RNS
 
 | |
3RHI
 
 | | DNA-binding protein HU from Bacillus anthracis | | Descriptor: | DNA-binding protein HU | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | DNA-binding protein HU from Bacillus anthracis.
To be Published
|
|
3Q7I
 
 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with 6-phosphogluconic acid. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-PHOSPHOGLUCONIC ACID, CALCIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3OMT
 
 | | Putative antitoxin component, CHU_2935 protein, from Xre family from Prevotella buccae. | | Descriptor: | PHOSPHATE ION, uncharacterized protein | | Authors: | Osipiuk, J, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystal structure of putative antitoxin component, CHU_2935 protein, from Xre family from Prevotella buccae.
To be Published
|
|
3Q88
 
 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with ribose 1,5-bisphosphate. | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3E6Q
 
 | | Putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure of putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa.
To be Published
|
|
3EY5
 
 | | Putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron. | | Descriptor: | Acetyltransferase-like, GNAT family, SULFATE ION | | Authors: | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-17 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystal structure of putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron.
To be Published
|
|
3FOV
 
 | | Crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris | | Descriptor: | NITRATE ION, UPF0102 protein RPA0323 | | Authors: | Osipiuk, J, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-02 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris.
To be Published
|
|
3CO5
 
 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | Descriptor: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | Authors: | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
3CWC
 
 | | Crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative glycerate kinase 2 | | Authors: | Osipiuk, J, Zhou, M, Holzle, D, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-08 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | X-ray crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2.
To be Published
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
3CNG
 
 | | Crystal structure of NUDIX hydrolase from Nitrosomonas europaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
3DV9
 
 | | Putative beta-phosphoglucomutase from Bacteroides vulgatus. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-18 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | X-ray crystal structure of putative beta-phosphoglucomutase from Bacteroides vulgatus.
To be Published
|
|
