6IN9
 
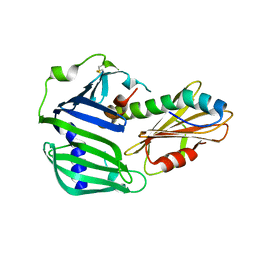 | | Crystal structure of MucB in complex with MucA(peri) | | Descriptor: | Sigma factor AlgU negative regulatory protein, Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6INB
 
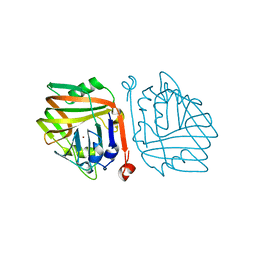 | |
6INC
 
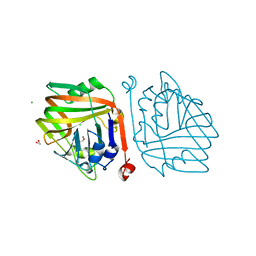 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6IN7
 
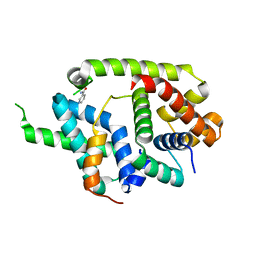 | | Crystal structure of AlgU in complex with MucA(cyto) | | Descriptor: | NICOTINAMIDE, RNA polymerase sigma-H factor, Sigma factor AlgU negative regulatory protein | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
4XHD
 
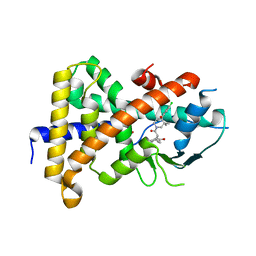 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN WITH COMPOUND-1 | | Descriptor: | GLYCEROL, N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-2-cyclopropylacetamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Khan, J.A, Camac, D.M. | | Deposit date: | 2015-01-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Developing Adnectins That Target SRC Co-Activator Binding to PXR: A Structural Approach toward Understanding Promiscuity of PXR.
J.Mol.Biol., 427, 2015
|
|
7D9F
 
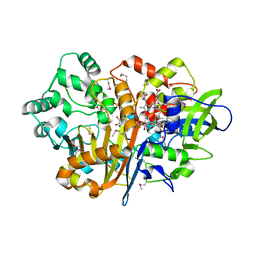 | | SpdH Spermidine dehydrogenase SeMet Structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9J
 
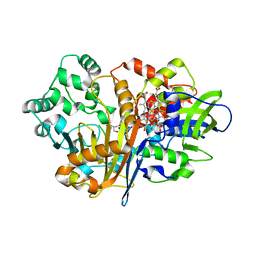 | | SpdH Spermidine dehydrogenase Y443A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9I
 
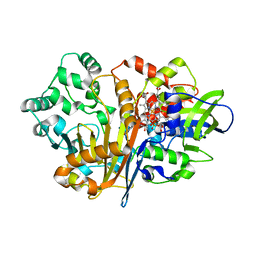 | | SpdH Spermidine dehydrogenase D282A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, Spermidine dehydrogenase, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9H
 
 | | SpdH Spermidine dehydrogenase N33 truncation structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9G
 
 | | SpdH Spermidine dehydrogenase native structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, Spermidine dehydrogenase, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D3E
 
 | | Cryo-EM structure of human DUOX1-DUOXA1 in low-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
7E7G
 
 | |
7EF1
 
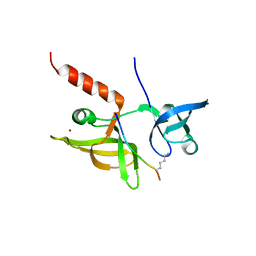 | |
7EF2
 
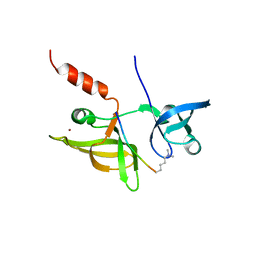 | |
7EF3
 
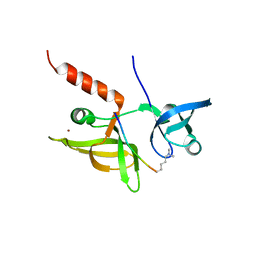 | |
7EEZ
 
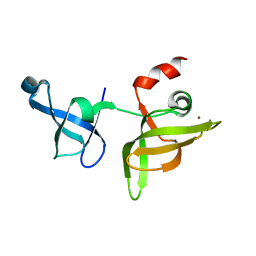 | | crystal structure of maize SHH2 SAWADEE domain | | Descriptor: | HB transcription factor, ZINC ION | | Authors: | Wang, Y, Du, J. | | Deposit date: | 2021-03-20 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Recognition of H3K9me1 by maize RNA-directed DNA methylation factor SHH2.
J Integr Plant Biol, 63, 2021
|
|
7EF0
 
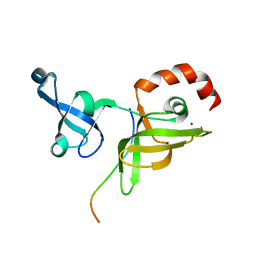 | |
7E53
 
 | |
7BVD
 
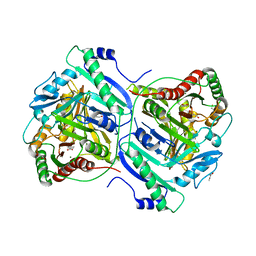 | | Anthranilate synthase component I (TrpE)[Mycolicibacterium smegmatis] | | Descriptor: | Anthranilate synthase component 1, BENZOIC ACID, GLYCEROL, ... | | Authors: | Chen, Y, Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit I of the anthranilate synthase complex of Mycolicibacterium smegmatis
Biochem.Biophys.Res.Commun., 527, 2020
|
|
7D3F
 
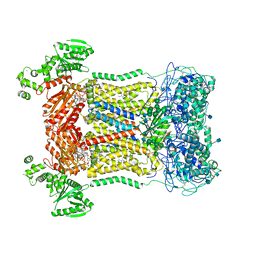 | | Cryo-EM structure of human DUOX1-DUOXA1 in high-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dual oxidase 1, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
7D54
 
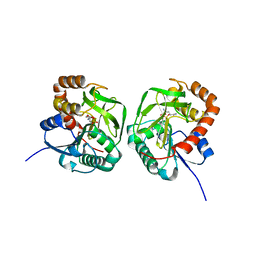 | | Crstal structure MsGATase with Gln | | Descriptor: | GLUTAMINE, Glutamine amidotransferase class-I | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D50
 
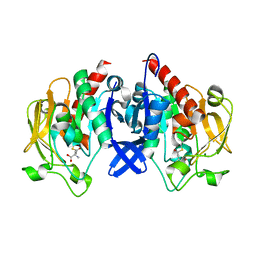 | | SpuA mutant - H221N with glutamyl-thioester | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D4R
 
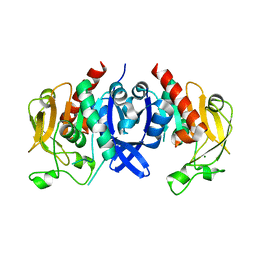 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D53
 
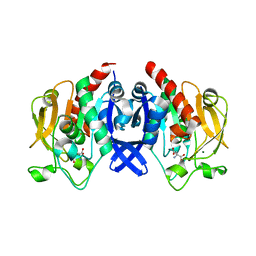 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7ET5
 
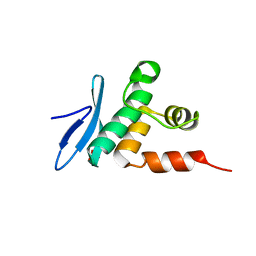 | | Crystal structure of Arabidopsis TEM1 AP2 domain | | Descriptor: | AP2/ERF and B3 domain-containing transcription repressor TEM1, SULFATE ION | | Authors: | Hu, H, Du, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
