3MKL
 
 | |
3L0Z
 
 | |
3M0Z
 
 | | Crystal structure of putative aldolase from Klebsiella pneumoniae. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative aldolase from Klebsiella pneumoniae.
To be Published
|
|
3M6Y
 
 | | Structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at 1.45 a resolution. | | Descriptor: | 4-Hydroxy-2-oxoglutarate aldolase, CALCIUM ION, CHLORIDE ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of 4-Hydroxy-2-Oxoglutarate Aldolase from Bacillus Cereus at 1.45 A Resolution.
To be Published
|
|
3MVN
 
 | | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP | | Descriptor: | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP
To be Published
|
|
3MUX
 
 | | The Crystal Structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus anthracis to 1.45A | | Descriptor: | CHLORIDE ION, SODIUM ION, putative 4-hydroxy-2-oxoglutarate aldolase | | Authors: | Stein, A.J, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-03 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus anthracis to 1.45A
To be Published
|
|
3MYF
 
 | |
3MZY
 
 | |
3MR7
 
 | |
3N2Q
 
 | |
3NE7
 
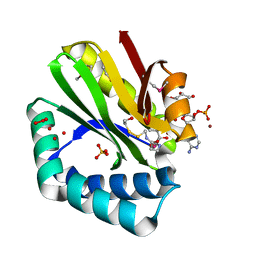 | | Crystal structure of paia n-acetyltransferase from thermoplasma acidophilum in complex with coenzyme a | | Descriptor: | ACETYLTRANSFERASE, BETA-MERCAPTOETHANOL, COENZYME A, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3BUU
 
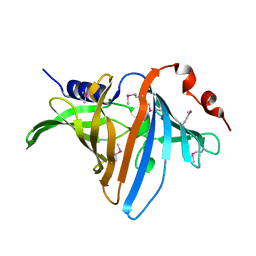 | |
3BV8
 
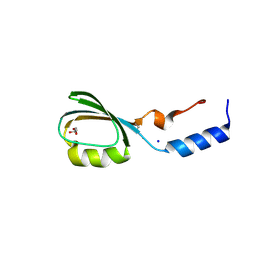 | | Crystal structure of the N-terminal domain of tetrahydrodipicolinate acetyltransferase from Staphylococcus aureus | | Descriptor: | GLYCEROL, SODIUM ION, Tetrahydrodipicolinate acetyltransferase | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the N-terminal domain of tetrahydrodipicolinate acetyltransferase from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3CP3
 
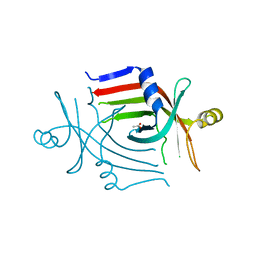 | |
3CCY
 
 | |
3CLQ
 
 | |
3D0J
 
 | | Crystal structure of conserved protein of unknown function CA_C3497 from Clostridium acetobutylicum ATCC 824 | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein CA_C3497 | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of Conserved Protein of Unknown Function CA_C3497 from Clostridium acetobutylicum ATCC 824.
To be Published
|
|
3D0F
 
 | | Structure of the BIG_1156.2 domain of putative penicillin-binding protein MrcA from Nitrosomonas europaea ATCC 19718 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Penicillin-binding 1 transmembrane protein MrcA | | Authors: | Cuff, M.E, Mulligan, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the BIG_1156.2 domain of putative penicillin-binding protein MrcA from Nitrosomonas europaea ATCC 19718.
TO BE PUBLISHED
|
|
3D3Y
 
 | | Crystal structure of a conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Uncharacterized protein | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-13 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of a conserved protein from Enterococcus faecalis V583.
To be Published
|
|
3E0Y
 
 | |
3CO5
 
 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | Descriptor: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | Authors: | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
3CJ8
 
 | | Crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Enterococcus faecalis V583 | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-acetyltransferase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Enterococcus faecalis V583.
To be Published
|
|
3BZ6
 
 | |
3DNX
 
 | |
3CQB
 
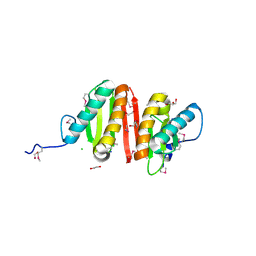 | | Crystal structure of heat shock protein HtpX domain from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-02 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The crystal structure of heat shock protein HtpX domain from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
