6UDI
 
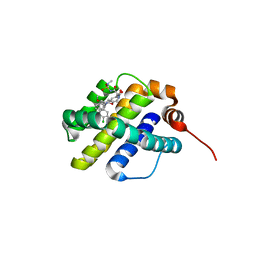 | | X-ray co-crystal structure of compound 20 with Mcl-1 | | Descriptor: | (4S,7aR,9aR,10S,11E,18R)-6'-chloro-N-(dimethylsulfamoyl)-18-hydroxy-10-methoxy-15-methyl-16-oxo-3',4',7,7a,8,9,9a,10,13,14,15,16,17,18-tetradecahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)cyclobuta[n][1,4]oxazepino[4,3-a][1,8]diazacyclohexadecine-4,1'-naphthalene]-18-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X, Whittington, D. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
8AOL
 
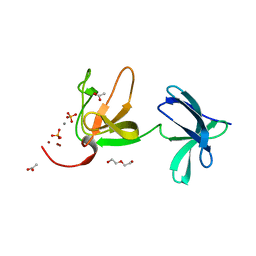 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain III (aa 363-499) | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sagmeister, T, Damisch, E, Eder, M, Dordic, A, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8AON
 
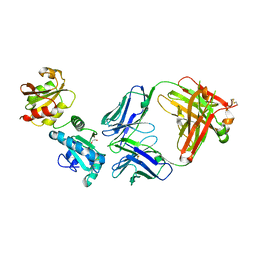 | |
8AQF
 
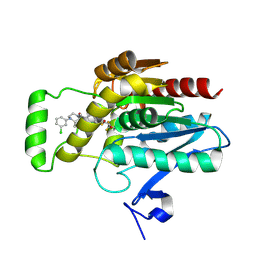 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND LEI-515 | | Descriptor: | 1-[(~{R})-[2-chloranyl-4-[(2~{S},3~{S})-4-(3-chlorophenyl)-2,3-dimethyl-piperazin-1-yl]carbonyl-phenyl]sulfinyl]-3,3-bis(fluoranyl)pentan-2-one, Monoglyceride lipase | | Authors: | Jiang, M, Huizenga, M, Wirt, J, Paloczi, J, Amedi, A, van der Berg, R, Benz, J, Collin, L, Deng, H, Driever, W, Florea, B, Grether, U, Janssen, A, Heitman, L, Lam, T.W, Mohr, F, Pavlovic, A, Ruf, I, Rutjes, H, Stevens, F, van der Vliet, D, van der Wel, T, Wittwer, M, Boeckel, C, Pacher, P, Hohmann, A, van der Stelt, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A monoacylglycerol lipase inhibitor showing therapeutic efficacy in mice without central side effects or dependence.
Nat Commun, 14, 2023
|
|
6P0M
 
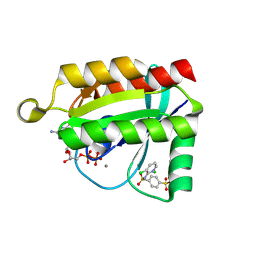 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(3-chloropyridin-2-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5K7Q
 
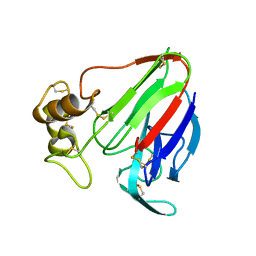 | | MicroED structure of thaumatin at 2.5 A resolution | | Descriptor: | Thaumatin-1 | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
8A2O
 
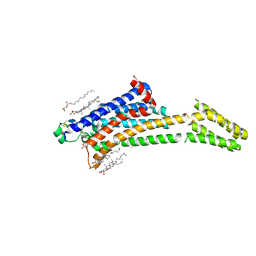 | | Room-temperature structure of the stabilised A2A-Theophylline complex determined by synchrotron serial crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Moraes, I, Kwan, T.O.C, Axford, D. | | Deposit date: | 2022-06-06 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | A versatile approach to high-density microcrystals in lipidic cubic phase for room-temperature serial crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
8A2P
 
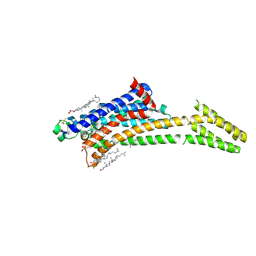 | | Room-temperature structure of the stabilised A2A-LUAA47070 complex determined by synchrotron serial crystallography | | Descriptor: | 4-(3,3-dimethylbutanoylamino)-3,5-bis(fluoranyl)-~{N}-(1,3-thiazol-2-yl)benzamide, Adenosine receptor A2a,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Moraes, I, Kwan, T.O.C, Axford, D. | | Deposit date: | 2022-06-06 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A versatile approach to high-density microcrystals in lipidic cubic phase for room-temperature serial crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
8ALU
 
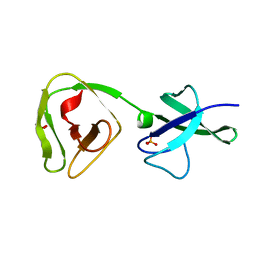 | | Crystal structure of the teichoic acid binding domain of SlpA, S-layer protein from Lactobacillus acidophilus (aa. 314-444) | | Descriptor: | PHOSPHATE ION, S-layer protein | | Authors: | Eder, M, Dordic, A, Sagmeister, T, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6TX7
 
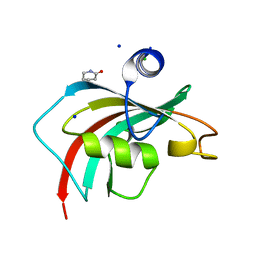 | |
8AJZ
 
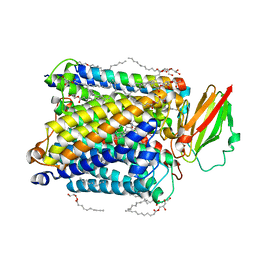 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase at 2 milliseconds after irradiation by a 532 nm laser | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Bosman, R, Dahl, P, Nango, E, Tanaka, R, Zoric, D, Svensson, E, Nakane, T, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
3DKT
 
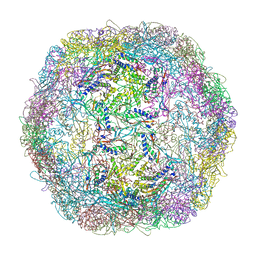 | | Crystal structure of Thermotoga maritima encapsulin | | Descriptor: | Maritimacin, Putative uncharacterized protein | | Authors: | Sutter, M, Boehringer, D, Gutmann, S, Weber-Ban, E, Ban, N. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis of enzyme encapsulation into a bacterial nanocompartment
Nat.Struct.Mol.Biol., 15, 2008
|
|
5K7T
 
 | | MicroED structure of thermolysin at 2.5 A resolution | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
7PTF
 
 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with novobiocin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B, ... | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PTG
 
 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
5K7O
 
 | | MicroED structure of lysozyme at 1.8 A resolution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.8 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
8AFQ
 
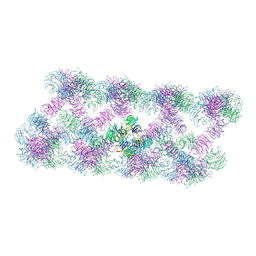 | | Tube assembly of Atg18-PR72AA | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFW
 
 | | Tube assembly of Atg18-WT | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFY
 
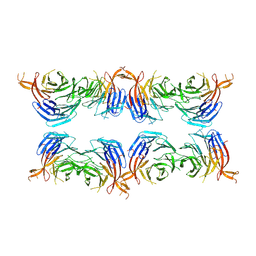 | | Subtomogram average of membrane-bound Atg18 oligomers | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AHY
 
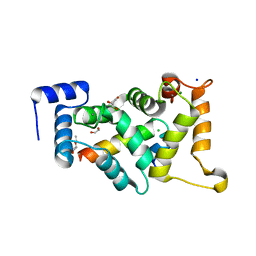 | |
8ALH
 
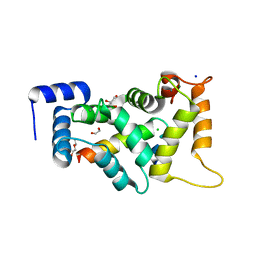 | |
8A8F
 
 | | Crystal structure of Glc7 phosphatase in complex with the regulatory region of Ref2 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, RNA end formation protein 2, ... | | Authors: | Carminati, M, Manav, C.M, Bellini, D, Passmore, L.A. | | Deposit date: | 2022-06-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A direct interaction between CPF and RNA Pol II links RNA 3' end processing to transcription.
Mol.Cell, 83, 2023
|
|
7PQI
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQM
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
8ALM
 
 | |
