3BOF
 
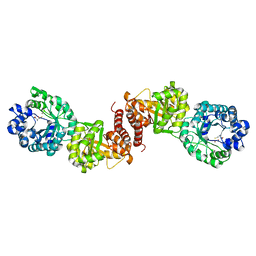 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ and Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BOL
 
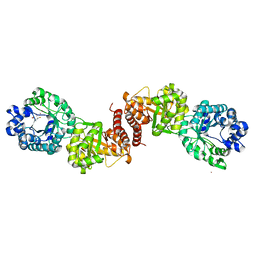 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BQ6
 
 | |
3QIT
 
 | |
3IV9
 
 | |
3KG6
 
 | |
3KG8
 
 | |
3IVA
 
 | | Structure of the B12-dependent Methionine Synthase (MetH) C-teminal half with AdoHcy bound | | Descriptor: | COBALAMIN, Methionine synthase, NITRATE ION, ... | | Authors: | Pattridge, K.A, Koutmos, M, Smith, J.L. | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the reactivation of cobalamin-dependent methionine synthase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KG9
 
 | |
3KG7
 
 | |
3LCR
 
 | | Thioesterase from Tautomycetin Biosynthhetic Pathway | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Tautomycetin biosynthetic PKS | | Authors: | Akey, D.L, Scaglione, J.B, Smith, J.L, Sherman, D.H. | | Deposit date: | 2010-01-11 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the tautomycetin thioesterase: analysis of a stereoselective polyketide hydrolase.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3NNJ
 
 | | Halogenase domain from CurA module (apo Hal) | | Descriptor: | CurA | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NNL
 
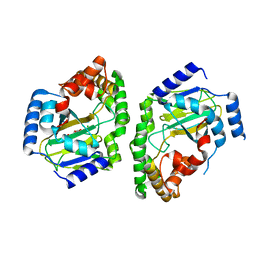 | | Halogenase domain from CurA module (crystal form III) | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, CurA, ... | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NNF
 
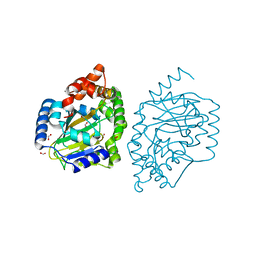 | | Halogenase domain from CurA module with Fe, chloride, and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, CurA, ... | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3SBY
 
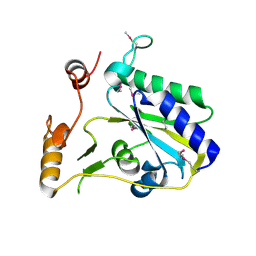 | | Crystal Structure of SeMet-Substituted Apo-MMACHC (1-244), a human B12 processing enzyme | | Descriptor: | Methylmalonic aciduria and homocystinuria type C protein | | Authors: | Koutmos, M, Gherasim, C, Smith, J.L, Banerjee, R. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural basis of multifunctionality in a vitamin B12-processing enzyme.
J.Biol.Chem., 286, 2011
|
|
3PC4
 
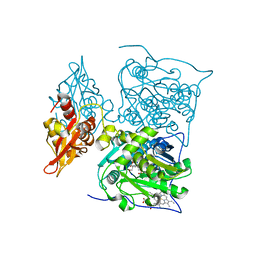 | |
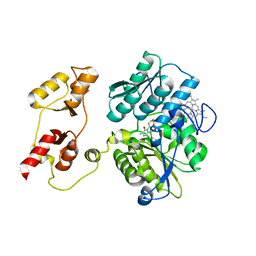
3PC2
 
 | |
3QMV
 
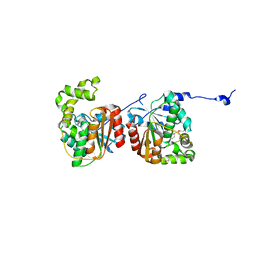 | |
3SSO
 
 | |
3NNM
 
 | |
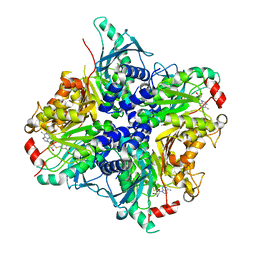
3SSM
 
 | |
3SSN
 
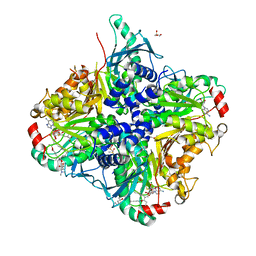 | | MycE Methyltransferase from the Mycinamycin Biosynthetic Pathway in Complex with Mg, SAH, and Mycinamycin VI | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A new structural form in the SAM/metal-dependent o‑methyltransferase family: MycE from the mycinamicin biosynthetic pathway.
J.Mol.Biol., 413, 2011
|
|
3U1T
 
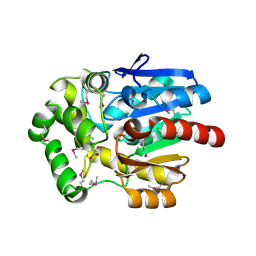 | | Haloalkane Dehalogenase, DmmA, of marine microbial origin | | Descriptor: | CHLORIDE ION, DmmA Haloalkane Dehalogenase, MALONATE ION | | Authors: | Gehret, J.J, Smith, J.L. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of DmmA, a marine haloalkane dehalogenase.
Protein Sci., 21, 2012
|
|
3QMW
 
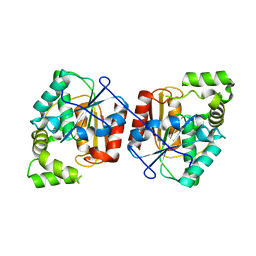 | | RedJ with PEG molecule bound in the active site | | Descriptor: | TETRAETHYLENE GLYCOL, Thioesterase | | Authors: | Whicher, J.R, Smith, J.L. | | Deposit date: | 2011-02-05 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Function of the RedJ Protein, a Thioesterase from the Prodiginine Biosynthetic Pathway in Streptomyces coelicolor.
J.Biol.Chem., 286, 2011
|
|
3SC0
 
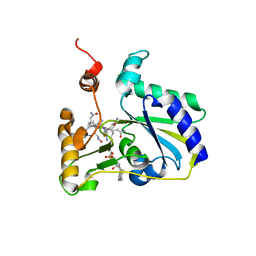 | | Crystal Structure of MMACHC (1-238), a human B12 processing enzyme, complexed with MethylCobalamin | | Descriptor: | CO-METHYLCOBALAMIN, Methylmalonic aciduria and homocystinuria type C protein | | Authors: | Koutmos, M, Gherasim, C, Smith, J.L, Banerjee, R. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of multifunctionality in a vitamin B12-processing enzyme.
J.Biol.Chem., 286, 2011
|
|
