8CMR
 
 | | Linear specific OTU-type DUB SnOTU from the pathogen S. negenvensis in complex with linear di-ubiquitin | | Descriptor: | OTU domain-containing protein, Polyubiquitin-B | | Authors: | Uthoff, M, Hermanns, T, Boll, V, Hofmann, K, Baumann, U. | | Deposit date: | 2023-02-21 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Functional and structural diversity in deubiquitinases of the Chlamydia-like bacterium Simkania negevensis.
Nat Commun, 14, 2023
|
|
7ODO
 
 | |
7ODQ
 
 | |
7ODP
 
 | |
7OEY
 
 | | Neisseria gonnorhoeae variant E93Q at 1.35 angstrom resolution | | Descriptor: | GLYCEROL, SODIUM ION, SUCCINIC ACID, ... | | Authors: | Rabe von Pappenheim, F, Wensien, M, Tittmann, K. | | Deposit date: | 2021-05-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Widespread occurrence of covalent lysine-cysteine redox switches in proteins.
Nat.Chem.Biol., 18, 2022
|
|
7OQM
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.05 Angstroms resolution, 20 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQN
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 30 minutes soaking | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQF
 
 | | Human OMPD-domain of UMPS in complex with OMP at 1.05 Angstrom resolution, 5 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQI
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.15 Angstrom resolution, 10 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQK
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 15 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OUZ
 
 | |
7OTU
 
 | | Human OMPD-domain of UMPS in complex with 6-hydroxy-UMP at 0.95 Angstroms resolution, crystal 2 | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Isoform 2 of Uridine 5'-monophosphate synthase | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
3ZGD
 
 | | crystal structure of a KEAP1 mutant | | Descriptor: | ACETATE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1, SODIUM ION | | Authors: | Hoerer, S, Reinert, D, Ostmann, K, Hoevels, Y, Nar, H. | | Deposit date: | 2012-12-17 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal-Contact Engineering to Obtain a Crystal Form of the Kelch Domain of Human Keap1 Suitable for Ligand-Soaking Experiments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3ZGC
 
 | | crystal structure of the KEAP1-NEH2 complex | | Descriptor: | ACETATE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1, NUCLEAR FACTOR ERYTHROID 2-RELATED FACTOR 2 | | Authors: | Hoerer, S, Reinert, D, Ostmann, K, Hoevels, Y, Nar, H. | | Deposit date: | 2012-12-17 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal-Contact Engineering to Obtain a Crystal Form of the Kelch Domain of Human Keap1 Suitable for Ligand-Soaking Experiments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4AE4
 
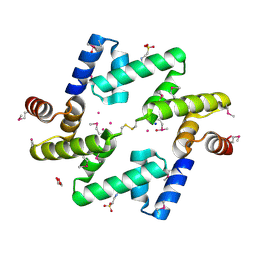 | | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a novel SOUBA domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Agromayor, M, Soler, N, Caballe, A, Kueck, T, Freund, S.M, Allen, M.D, Bycroft, M, Perisic, O, Ye, Y, McDonald, B, Scheel, H, Hofmann, K, Neil, S.J.D, Martin-Serrano, J, Williams, R.L. | | Deposit date: | 2012-01-06 | | Release date: | 2012-03-21 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain.
Structure, 20, 2012
|
|
4AXA
 
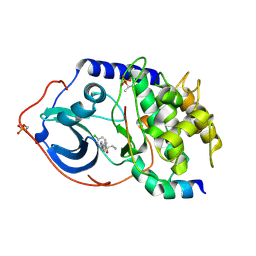 | | Structure of PKA-PKB chimera complexed with (1S)-2-amino-1-(4- chlorophenyl)-1-(4-(1H-pyrazol-4-yl)phenyl)ethan-1-ol | | Descriptor: | (2S)-2-(4-chlorophenyl)-2-hydroxy-2-[4-(1H-pyrazol-4-yl)phenyl]ethanaminium, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Davies, T.G, Yap, T.A, Walton, M.I, Grimshaw, K.M, tePoele, R.H, Eve, P.D, Valenti, M.R, deHavenBrandon, A.K, Martins, V, Zetterlund, A, Heaton, S.P, Heinzmann, K, Jones, P.S, Feltell, R.E, Reule, M, Woodhead, S.J, Lyons, J.F, Raynaud, F.I, Eccles, S.A, Workman, P, Thompson, N.T, Garrett, M.D. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-25 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | At13148 is a Novel, Oral Multi-Agc Kinase Inhibitor with Potent Pharmacodynamic and Antitumor Activity.
Clin.Cancer Res., 18, 2012
|
|
6FFL
 
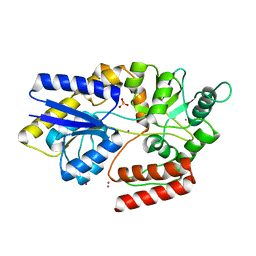 | | Maltose/maltodextrin-binding domain MalE from Bdellovibrio bacteriovorus bound to maltotriose | | Descriptor: | Maltose/maltodextrin transport permease homologue, PLATINUM (II) ION, SULFATE ION, ... | | Authors: | Licht, A, Werther, T, Bommer, M, Neumann, K, Schneider, E. | | Deposit date: | 2018-01-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Structural and functional characterization of a maltose/maltodextrin ABC transporter comprising a single solute binding domain (MalE) fused to the transmembrane subunit MalF.
Res. Microbiol., 170, 2018
|
|
5HHT
 
 | |
6H60
 
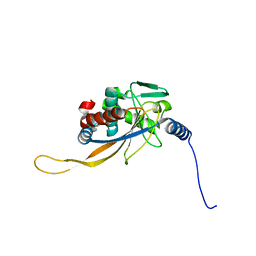 | | pseudo-atomic structural model of the E3BP component of the human pyruvate dehydrogenase multienzyme complex | | Descriptor: | Pyruvate dehydrogenase protein X component, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
6H55
 
 | | core of the human pyruvate dehydrogenase (E2) | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
8RPI
 
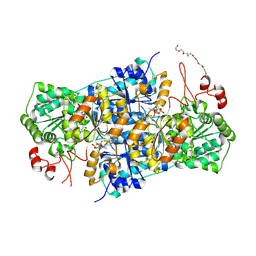 | | JanthE from Janthinobacterium sp. HH01, lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RPH
 
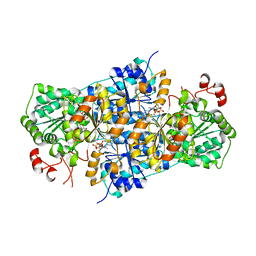 | | JanthE from Janthinobacterium sp. HH01,ketobutyryl-ThDP | | Descriptor: | (2~{S})-2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-2-yl]-2-oxidanyl-butanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8WA9
 
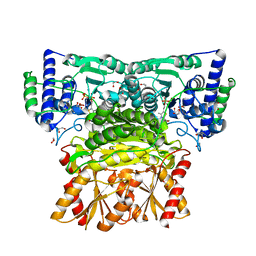 | | Human transketolase soaked with donor ketose D-fructose | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Liu, Z, Tittmann, K, Dai, S. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8WA7
 
 | | E.coli transketolase soaked with donor ketose D-fructose | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, ... | | Authors: | Liu, Z, Dai, S, Tittmann, K. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8WAA
 
 | | Human transketolase soaked with donor ketose D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Liu, Z, Dai, S, Tittmann, K. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
