5TIE
 
 | | x-ray structure of acyl-CoA thioesterase I, TesA, mutant M141L/Y145K/L146K at pH 7.5 in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
1NS4
 
 | |
1NSX
 
 | |
5U21
 
 | | X-ray structure of the WlaRF aminotransferase from Campylobacter jejuni, K184A mutant in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
1NSV
 
 | |
1NS0
 
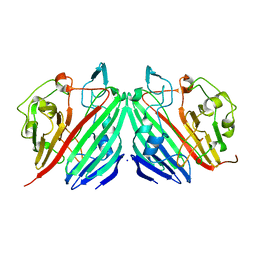 | |
1NS7
 
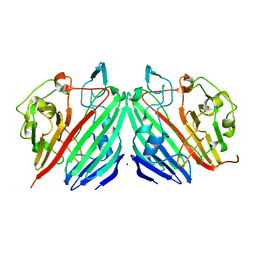 | |
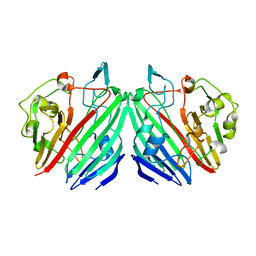
5TIF
 
 | | x-ray structure of acyl-CoA thioesterase I, TesA, triple mutant M141L/Y145K/L146K in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, DI(HYDROXYETHYL)ETHER, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
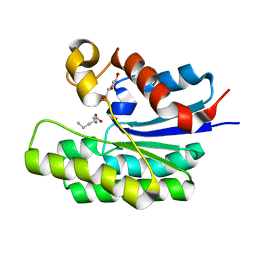
1L7J
 
 | |
1L7K
 
 | |
1KVU
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1J7A
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN D68K MUTANT | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, VanHooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollen, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1J7B
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN MUTANT E94K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, Vanhooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollin, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
3M9V
 
 | |
6OFU
 
 | | X-ray crystal structure of the YdjI aldolase from Escherichia coli K12 | | Descriptor: | CHLORIDE ION, YdjI aldolase, ZINC ION | | Authors: | Dopkins, B.J, Thoden, J.B, Huddleston, J.P, Narindoshvili, T, Fose, B, Rachel, F.M, Holden, H.M. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Characterization of YdjI, an Aldolase of Unknown Specificity inEscherichia coliK12.
Biochemistry, 58, 2019
|
|
6Q03
 
 | | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine) | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine)
To Be Published
|
|
6Q0A
 
 | | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116D, n the presence of UDP-N-acetylmuramic acid
To Be Published
|
|
6Q11
 
 | | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID
To Be Published
|
|
6Q0Y
 
 | | Crystal structure of MurA from Clostridium difficile, mutant C116S, in the presence of Uridine-Diphosphate-N-Acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutant C116S, in the presence of Uridine-Diphosphate-N-Acetylglucosamine
To Be Published
|
|
6PZ2
 
 | |
6NBP
 
 | | Crystal Structure of a Sugar N-Formyltransferase from the Plant Pathogen Pantoea ananatis | | Descriptor: | N-formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Hofmeister, D.L, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Investigation of a sugar N-formyltransferase from the plant pathogen Pantoea ananatis.
Protein Sci., 28, 2019
|
|
9BGP
 
 | | X-ray structure of the aminotransferase from Vibrio vulnificus responsible for the biosynthesis of 2,3-diacetamido-4-amino-2,3,4-trideoxy-arabinose in the presence of its internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, aminotransferase | | Authors: | Fait, D.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical Investigation of an Aminotransferase Required for the Production of 2,3,4-triacetamido-2,3,4-trideoxy-L-arabinose
To Be Published
|
|
9BGR
 
 | | X-ray structure of the aminotransferase from Vibrio vulnificus responsible for the biosynthesis of 2,3-diacetamido-4-amino-2,3,4-trideoxy-arabinose in the presence of its external aldimine with 2,3-diacetamido-4-amino-2,3,4-trideoxy-l-arabinose | | Descriptor: | (2R,3R,4R,5R)-3,4-diacetamido-5-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methoxy)oxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Fait, D.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Biochemical Investigation of an Aminotransferase Required for the Production of 2,3,4-triacetamido-2,3,4-trideoxy-L-arabinose
To Be Published
|
|
9CJ0
 
 | |
5KTA
 
 | | Apo FdhC- a nucleotide-linked sugar GNAT | | Descriptor: | DIMETHYL SULFOXIDE, FdhC, SULFATE ION | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
