7X8Q
 
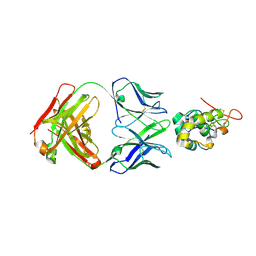 | | Frizzled-10 CRD in complex with F10_A9 Fab | | Descriptor: | Antibody F10_A9 Fab, Heavy chain, Light chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
7X8T
 
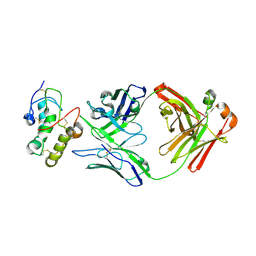 | | Frizzled 10 CRD in complex with hB9L9.3 Fab | | Descriptor: | Antibody hB9L9.3 Fab, Heavy chain, Light chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
7X8P
 
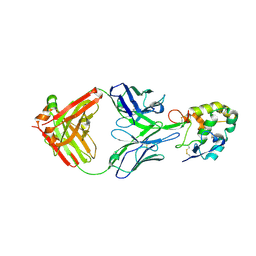 | | Frizzled 2 CRD in complex with pF7_A5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody pF7_A5 Fab, Heavy chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
6SGF
 
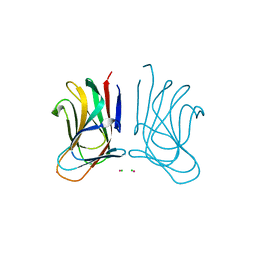 | | Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. | | Descriptor: | Beta-xylanase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Leth, M.L, Guskov, A, Slotboom, D. | | Deposit date: | 2019-08-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Molecular insight into a new low-affinity xylan binding module from the xylanolytic gut symbiont Roseburia intestinalis.
Febs J., 287, 2020
|
|
7VU6
 
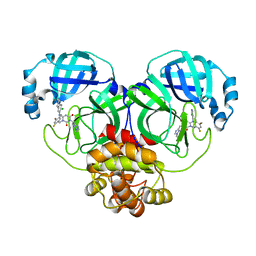 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 3 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Yamamoto, S, Yamane, J, Tachibana, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
7VTH
 
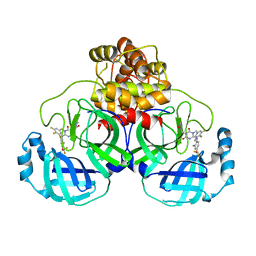 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 2-[4-[[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]amino]-2,6-bis(oxidanylidene)-3-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazin-1-yl]-N-methyl-ethanamide, 3C-like proteinase | | Authors: | Yamamoto, S, Tachibana, Y. | | Deposit date: | 2021-10-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
2RVA
 
 | |
2RV9
 
 | |
3VJT
 
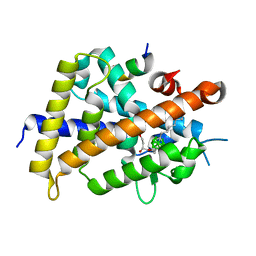 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(R)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
7DV6
 
 | | Discovery of Functionally Selective Transforming Growth Factor beta Type II Receptor (TGF-beta RII) Inhibitors as Anti-Fibrosis Agents | | Descriptor: | 5-[(3S)-5,5-dimethyloxolan-3-yl]-6-methoxy-3-(2-methoxypyridin-4-yl)pyrazolo[1,5-a]pyrimidine, TGF-beta receptor type-2 | | Authors: | Nishihata, J, Nomura, A, Miwa, S, Doi, S, Adachi, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of Selective Transforming Growth Factor beta Type II Receptor Inhibitors as Antifibrosis Agents.
Acs Med.Chem.Lett., 12, 2021
|
|
3VJS
 
 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(S)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
7D83
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-(2-(3-cyclohexylureido)-3,6-dimethyl-5-(5-methylchroman-6-yl)pyridin-4-yl)acetic acid | | Descriptor: | (2S)-2-[2-(cyclohexylcarbamoylamino)-3,6-dimethyl-5-(5-methyl-3,4-dihydro-2H-chromen-6-yl)pyridin-4-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase, SULFATE ION | | Authors: | Sugiyama, S, Sekiguchi, Y. | | Deposit date: | 2020-10-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of novel HIV-1 integrase-LEDGF/p75 allosteric inhibitors based on a pyridine scaffold forming an intramolecular hydrogen bond.
Bioorg.Med.Chem.Lett., 33, 2020
|
|
8ZM2
 
 | | Structure of human pyruvate dehydrogenase kinase 2 complexed with compound 16 | | Descriptor: | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial, methyl (9~{R})-9-oxidanyl-9-(trifluoromethyl)fluorene-4-carboxylate | | Authors: | Akai, S, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-22 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design and synthesis of novel fluorene derivatives as inhibitors of pyruvate dehydrogenase kinase.
Bioorg.Med.Chem.Lett., 109, 2024
|
|
8ZM1
 
 | | Structure of human pyruvate dehydrogenase kinase 2 complexed with compound 6 | | Descriptor: | (5~{R})-5-propan-2-ylindeno[1,2-b]pyridin-5-ol, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial | | Authors: | Akai, S, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-22 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Design and synthesis of novel fluorene derivatives as inhibitors of pyruvate dehydrogenase kinase.
Bioorg.Med.Chem.Lett., 109, 2024
|
|
7X56
 
 | | A CBg-ParM filament with ADP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ParM/StbA family protein | | Authors: | Koh, A, Ali, S, Robinson, R, Narita, A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7X59
 
 | | A CBg-ParM filament with GTP or GDPPi | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, ParM/StbA family protein | | Authors: | Koh, A, Ali, S, Robinson, R, Narita, A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7X54
 
 | | A CBg-ParM filament with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ParM/StbA family protein | | Authors: | Koh, A, Ali, S, Robinson, R, Narita, A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7X55
 
 | |
